
Genomic profiling of extracellular vesicle-derived DNA from bronchoalveolar lavage fluid of patients with lung adenocarcinoma
Introduction
Next-generation sequencing (NGS) of DNA from tumor tissues of patients with non-small cell lung cancer (NSCLC) is recommended by National Comprehensive Cancer Network (NCCN) for identifying therapeutically relevant cancer genome alterations and facilitating appropriate counseling for clinical trials (1). However, NGS for tumor molecular genotyping in advanced lung cancer sometimes presents practical challenges such as the availability of adequate tissue biopsy specimens, the need for repeated biopsies after progression, and intratumoral heterogeneity. About 30% of tumor samples yields insufficient or inadequate tissue for molecular analysis (2,3).
Liquid biopsy using cell-free circulating tumor DNA (ctDNA) has emerged as an attractive practical alternative to tissue biopsy for overcoming these practical issues in clinical practice. A variety of epidermal growth factor receptor (EGFR) mutation detection assays, such as droplet digital PCR (ddPCR), BEAMing (beads, emulsion, amplification, and magnetics), and peptide nucleic acid (PNA)-mediated polymerase chain reaction (PCR) clamping, have been used to detect recurrent mutations in specific genes such as EGFR and KRAS using ctDNA of NSCLC patients (4-6). Furthermore, NGS-based liquid biopsy was recently used for the comprehensive genomic profiling of NSCLC using plasma ctDNA (7,8). However, its clinical application is limited due to the low sensitivity, unstable nature, and high degradation rate of ctDNA and the high admixture of normal DNA in ctDNA (9,10).
To resolve these limitations, we have developed a novel strategy for EGFR genotyping using DNA derived from extracellular vesicles (EVs) in bronchoalveolar lavage fluid (BALF) and pleural effusion (11,12). EVs are nanometer- to micrometer-scale, double-layer phospholipid membrane-bound particles released by almost all types of cells (13,14). EVs have been shown to contain various bioactive molecules such as proteins, RNA, and DNA, and they are released abundantly by tumor cells (14,15). Several recent studies on various cancers have reported the potential usefulness of EV-derived DNA (EV DNA) as a liquid biopsy biosource for molecular analysis (16-18). Other studies also have shown that unlike the extensively fragmented nature of circulating cell-free DNA (cfDNA), the majority of DNA derived from the tumor EVs of cancer cell lines and NSCLC patients are long double-stranded and stable (11,12,19,20). This finding demonstrated the usefulness of EV DNA as a circulating diagnostic biomarker for cancer.
In this study, we investigated the reliability of BALF-EV as a source for DNA of sufficient quality and at adequate quantities for use in NGS analysis for the detection of somatic mutations in EGFR-mutated lung adenocarcinoma in comparison with that of tissue DNA. To this end, we performed targeted NGS of DNA derived from BALF-EV (BALF EV DNA) of 20 patients with EGFR-mutated lung adenocarcinoma and DNA from matched formalin-fixed paraffin-embedded (FFPE) tissue samples. We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tlcr-20-888).
Methods
Patients
In this study, a total of 20 patients with lung adenocarcinoma were enrolled. Eligible patients had histologically confirmed adenocarcinoma. Clinicopathological information, including age, sex, smoking history, and stage of cancer, was obtained retrospectively by reviewing medical records. The pathologic stage of cancer was defined using the American Joint Committee on Cancer (AJCC) manual, eighth edition. This study was conducted in accordance with the amended Declaration of Helsinki (as revised in 2013). The study protocol was approved by the institutional review board of Konkuk University Medical Center (KUH1010899), and written informed consents were obtained from all patients.
Isolation of EVs and extraction of EV DNA
Bronchoalveolar lavage (BAL) was always performed before a biopsy to prevent contamination from possible bleeding after the bronchoscopic biopsy. A sample of 5 ml BALF was used for the isolation of EVs. Cells and debris were removed by centrifugation at 1,000 g for 10 min at 4 °C. Cell free BALF was spun in an ultracentrifuge tube at 200,000 g for 1 hour at 4 °C using a Beckman rotor (Beckman Coulter, Brea, CA, USA). The size of purified EV was analyzed using Nanosight NS300 (Malvern Instruments, Worcestershire, UK). The supernatant was carefully removed and the pellet was resuspended in 200 µL of PBS. EVs were lysed using the lysis buffer (10 mM Tris-HCl, 20% Triton X-100) and the DNA from lysed EVs was purified using a High-Pure PCR Template Preparation Kit (Roche Diagnostics, Mannheim, Germany). DNA concentration was assessed using a Quant-iTTM PicoGreen® dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA) on a QuantusTM Fluorometer (Promega, Madison, WI, USA). The quality and length of the purified DNA were analyzed using a 2200 Tapestion and Genomic DNA ScreenTape (Agilent Technologies, Santa Clara, CA, USA).
Negative-stain transmission electron microscopy (TEM)
The EV were visualized by negative stain TEM. For negative-stain TEM, purified EVs were fixed in 2% (vol/vol) paraformaldehyde for 5 min at room temperature. After fixation, 10 µL EV suspension was applied to formvar-/carbon-coated grids (200 mesh) for 1 min and was stained with 2% uranyl acetate. Excess uranyl formate was removed using a filter paper, and the grids were examined using a transmission electron microscope (H7600; Hitachi, Tokyo, Japan) at 80 kV. For the sections, EV pellets were fixed using 2.5% glutaraldehyde and 2% paraformaldehyde in sodium cacodylate buffer (pH 7.2) at 4 °C. Next, the samples were fixed again by using 1% osmium tetra-oxide for 30 min at 4 °C. The fixed samples were dehydrated using an ethanol series (50%, 60%, 70%, 80%, 90%, and 100% ethanol) for 20 min and were transferred to Spurr’s medium (Electron Microscopy Science, Hatfield, PA, USA). The samples were impregnated with and embedded into the same resin mixture, sectioned (60-nm-thick sections) with an ultramicrotome (Leica Ultracut UCT; Leica Microsystems, Vienna, Austria), and placed on nickel grids. The sections were stained with 2% uranyl acetate for 20 min and lead citrate for 10 min and were viewed under the transmission electron microscope.
EGFR genotyping
For detecting EGFR mutations and genotyping, a PANAMutyper™ R EGFR kit (Panagene, Daejeon, Korea) and CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA, USA) were used. All reactions had a total volume of 25 µL containing 70 ng of template DNA, the primer and PNA probe set along with a PCR master mix. PCR and the melting curve step were performed according to the manufacturer’s protocol. Fluorescence was measured on all four channels (FAM, ROX, Cy5, and HEX) (21,22).
DNA preparation, library generation and sequencing
Genomic DNA was extracted from fresh tissues using a QuickGene DNA tissue kit (KURABO, Osaka, Japan) and from FFPE tissues (five to ten 5 µm unstained FFPE slides) using a ReliaPrep FFPE gDNA Miniprep System (Promega).
DNA concentration was assessed using a Quant-iTTM PicoGreen® dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA) on a QuantusTM Fluorometer (Promega). The quality of DNA was measured using Genomic DNA ScreenTape on a 2200 TapeStation system (Agilent Technologies). Genomic DNA was fragmented to 150–200 bp using M220 Focused-ultrasonicator (Covaris, Woburn, MA, USA) followed by purification using 1.8x volumes of CeleMag clean-up beads (Celemics, Seoul, Korea). After the fragmentation, library preparation and target capture were performed using Agilent’s SureSelect XT target Enrichment kit and CancerSCANTM ver2.2 (Samsung Genome Institute, Seoul, Korea) respectively, according to the manufacturer’s recommendations (Agilent Technologies). We performed 8 cycles of PCR to generate the pre-capture library before hybrid selection and 12 cycles of PCR for amplification of the target-capture library with a barcode. The target-capture library was quantified using a Quant-iTTM PicoGreen® dsDNA Assay Kit (Invitrogen) on a QuantusTM Fluorometer (Promega). The quality of the target-capture library was analyzed using a High Sensitivity D1K ScreenTape on 2200 TapeStation system (Agilent Technologies) to determine the size distribution and to check for self-ligated adapter contamination. Based on the DNA concentration and average size of target-capture library, the library was normalized to a final concentration of 4 nM. After denaturing the library using 0.2 N NaOH, the library was diluted to 20 pM using hybridization buffer (HT1) (Illumina, San Diego, CA, USA). Seventy-five bp paired-end sequencing was performed on a NextSeq 500 platform according to the manufacturer’s instructions (Illumina).
Detection of variants
Four types of somatic variants (single nucleotide variations: SNVs, small insertions/deletions: small indels, copy number variations: CNVs and gene fusions) of BALF-EV or FFPE tissue samples were analyzed using CancerSCANTM ver2.2 (Samsung Genome Institute), a capture-based targeted sequencing platform (23). We used CancerSCANTM ver2.2 (Samsung Genome Institute) panel which targets 375 genes, covering about 2.5-megabase genomic regions including complete coding DNA sequences (CDSs) of 374 genes, selected intronic regions of 23 genes for fusion detection, and 1 kb TERT promoter region. In principle, variant analyses of NGS panel data were performed as described by Shin et al. (23) with minor modifications. Base call from sequencer was processed into FASTQ files using bcl2fastq (v2.18.0). The reads were aligned to a human reference genome (hg19) using BWA-MEM (v0.7.5). SAMtools (v0.1.18) and Picard (v1.93) were used for file conversion, read sorting, and duplicate removal. Local realignment and base quality recalibration were performed with GATK (v3.1). For detecting SNVs, the outputs from two variant callers, MuTect (v1.1.4) and LoFreq (v0.6.1), were combined to improve the sensitivity. Both callers were run with default parameters. Small indels were identified using Pindel (v0.2.5a4) with its default setting. Additional filters were applied to remove the following putative germline variants: (I) variants with high variant allele frequencies (VAF) (≥97%), except for the hotspot mutations; (II) variants with minor allele frequency (MAF) ≥3% in the >400 normal samples in our database; (III) variants with MAF ≥5% in public population databases such as ExAC (24), ESP6500 (25), and 1000Genome (26), and ethnic-specific databases including KRGDB (27) (n=1,100) and KOVA (28) (n=1,055); (IV) other frequently detected variants that are likely to be alignment artifact, as curated by manual review and compiled in our database. Putative somatic variants were functionally annotated using ANNOVAR (v2017-07-17) (29). The non-synonymous SNVs. (missense- and nonsense-SNVs), indel variants in CDS regions, splice site variants (SNVs. or indels at donor site +2 bp to acceptor site −2 bp), and variants in TERT promoter regions were reported. CNV and fusion variants were detected using in-house scripts (manuscript submitted) as described in by Shin et al. (23).
Tumor mutation burden (TMB) score was calculated as the total number of non-synonymous mutations identified divided by the megabase size of exonic regions covered by the CancerSCANTM ver2.2 (Samsung Genome Institute) panel. All synonymous SNVs, CNVs, or fusion variants were ignored for TMB scoring. Oncogenic driver mutations were included as described in by Samstein et al. (30).
Tumor purity estimation
The regions of copy number gain and loss were identified according to their adjusted coverage area relative to the copy number-neutral regions. The regions were then delineated, and tumor purity was inferred from the proportion of values which tumor clone estimated (23).
Statistical analysis
Categorical variables were summarized by calculating frequencies and percentages. Means, standard deviations, medians, and ranges, including minimal and maximal values, were used to determine numerical variables. For correlation statistics, the Spearman’s rank correlation test was used. All statistical analyses were carried out using SPSS Statistics version 25.0 (IBM Corp, Chicago, IL, USA), and a P value <0.05 was regarded as statistically significant.
Results
Clinicopathologic characteristics of patients
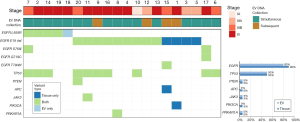
Clinical data of the 20 enrolled patients were reviewed and the clinicopathological characteristics are summarized in Tables 1,2. The majority of patients were at advanced stages of the disease. Their pathologic stages were as follows: 12 patients (60%), stage IV; 7 patients (35%), stage III, and 1 patient (5%), stage I. BALF cytology results yielded ten negatives, nine positives, and one atypical cell. All 20 patients had lung adenocarcinoma with at least one EGFR mutation, confirmed by EGFR genotyping of BALF EV DNA using PNA mediated PCR clamping method. The most common mutation, reported in 13 (65%) patients, was a deletion in exon 19, including one patient also with T790M mutation. Another 5 patients (25%) had a mutation in exon 21, L858R. One patient (5%) had mutations in both exons 18 and 20, and one patient (5%) had de novo T790M mutations. All patients except one had EGFR mutations in tumor tissues. The one patient with de novo T790M mutation identified by BALF EV-based EGFR genotyping had no EGFR mutation in the tissue EGFR genotyping. Four surgical type tissues (20%) and 16 biopsy type tissue (80%) were analyzed. All 20 patients were EGFR-tyrosine kinase inhibitor (TKI) naïve at the time of diagnosis and all but two patients received EGFR-TKI treatment after diagnosis.

Full table
Full table
Characterization of isolated BALF-EVs and EV DNA
EVs were isolated from the BALF by ultracentrifugation. These were observed with transmission electron microscopy (Figure S1) and the size and concentration were measured by nanosight (Figure S2). The average size and concentration of EVs from EGFR-TKI naïve patients (case 9) were 207.0±48.3 nm and (0.78±0.5)×1011 particles/mL, respectively (Table S1). Because isolated EVs from the BALF comprise of exosomes, microvesicles, apoptotic bodies, etc., the size range of EVs were heterogeneous, but predominantly concentrated between 100 and 300 nm (Figures S1,S2B). EV DNAs from the BALF existed in short and long sizes, but mostly in about 11 kb (Figure S3). The mean concentration of BALF EV DNA was 3.0±4.9 ng/µL (Table S2). DNase was not treated on EVs and therefore would contain DNAs from both vesicle surface and inside.
Assessment of NGS data quality
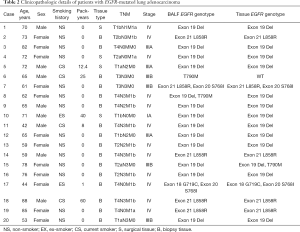
A comprehensive quality measurement was performed at every step (Figure S4). DNA obtained from the tumor tissue was matched with EV DNA in BALF. The median value of EV DNA yield extracted from BALF was 30.3 µg (range, 7.1–720 µg) and the median value of tissue DNA was 494 µg (range, 58–3,225 µg). The DNA yield from tissue samples was 100 times that from BALF-EVs (P=0.012, Figure 1A). In terms of sequencing statistics, median depth of coverage in tissue and BALF EV DNAs was 753× (range, 200×–1,117×) and 379× (range, 190×–755×), respectively (Figure 1B). The median sequencing uniformity in tissue and BALF EV DNAs was 97.1% (range, 49.5–98.2%) and 84.4% (range, 35.6–94.9%), respectively (Figure 1C), and the difference was statistically significant. Tumor purity was significantly lower in BALF EV DNA than tissue DNA (median 56% vs. 20%, Figure 1D). The median value of the estimated library size did not differ significantly between the two groups [46 G (range, 8–73 G) vs. 51 G (range, 34–68 G), P=0.45] (Figure 1E). However, the median fragment length of DNA was longer in BALF EV DNA than tissue DNA [175.5 bp (range, 160–186 bp) vs. 169.5 bp (range, 153–181 bp)] (Figure 1F).
Concordance of detected variants between tissue and BALF EV DNAs
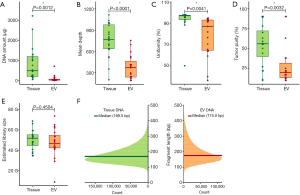
Each patient showed at least three alterations using the BALF EV DNA, with EGFR variants being most common, totalling detection of 580 alterations in 175 genes. NGS of BALF EV DNA detected 15 annotated and 66 known mutations. Nonsynonymous somatic mutations were identified with an overall concordance of 56% and clinically significant mutations in EGFR, TP53, PTEN, APC, JAK3, PIK3CA, and PRKAR1A, such as “actionable” genetic alterations, were identified with an overall concordance of 81% in in matched tissue and BALF EV DNAs (Figure 2A). When the somatic mutations including SNVs, insertions, or deletions identified in tissue DNA NGS was used as a reference, the by-variant sensitivity in BALF EV DNA was 62%, which increased to 83% when clinically significant somatic mutations identified in tissue DNA were used as a reference (Figure 2B).
Comparison of VAFs of clinically significant putative somatic mutations between tissue and BALF EV DNAs
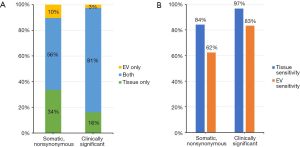
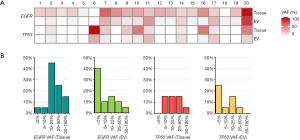
We assessed the correlation of the VAF of mutations identified in tissue and BALF EV DNAs. The median VAF in tissue and BALF EV DNAs were 19% (range, 0.12–97.6%) and 7.1% (range, 0.13–76.3%), respectively. The VAFs in clinically significant putative somatic variants detected in BALF EV DNA were significantly different from those in tissue DNA (P=0.016) (Figure 3A). A high degree of correlation was identified between the VAF in a tissue DNA and its matching BALF EV DNA, with an R2 value of 0.32 (Figure 3B).
Genomic profile comparison of lung adenocarcinoma tissue and BALF-EV
To assess the sensitivity and specificity of detected somatic mutations in BALF EV DNA, we compared the mutations identified in the NGS of tissue and BALF EV DNAs. Sensitivity for detection of the driver EGFR mutation was 80% (16/20) for BALF EV DNA NGS and 90% (18/20) for tissue DNA NGS (Figure 4). Four patients (case 1, 3, 5, and 15) had EGFR mutations in the tissue DNA NGS but none in the matching BALF EV DNA NGS. All discordant cases had exon 19 del mutation. One discordant case was identified at low allele frequency (AF) (<0.5%) in tissue DNA NGS. Conversely, only one patient (case 18) had no EGFR mutation in the tissue DNA NGS, but EGFR L858R mutation was identified in the BALF EV DNA NGS. One patient (case 6) had no EGFR T790M mutation in both BALF EV DNA NGS and tissue DNA NGS. There was no difference in the TNM stage and location of BAL between the concordant and discordant cases. The most frequent accompanying somatic mutation was TP53, which was mutated in 10 of 20 patients, with a concordance of 100% in tissue and BALF EV DNAs NGS (Figure 4). Few other somatic mutations were identified as EGFR alteration is the strongest driver mutation in lung adenocarcinoma. We compared the distribution of AFs for all EGFR and TP53 variants identified in tissue and BALF EV DNAs (Figure 5A). As expected, the most abundant AF in tissue DNA was in the range of 10–25% but <5% in the BALF EV DNA (Figure 5B).
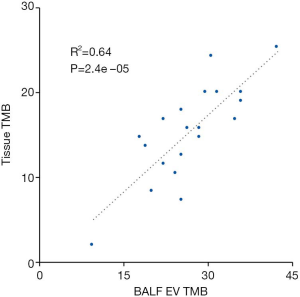
To assess the performance of BALF EV DNA in the TMB assay, we compared the results from a tissue TMB with the corresponding TMB results for the BALF EV DNA. TMB of tissue DNA algorithm included nonsynonymous mutations at an AF of ≥5%, whereas the TMB of BALF EV DNA algorithm included nonsynonymous mutations at an AF of ≥0.5%. Overall, the TMB scores in tissue and BALF EV DNAs showed a positive correlation (Spearman’s rank correlation =0.64, Figure 6).
Copy number gain was defined as the mean read depth of the gene divided by the median read depth of all autosome genes greater than 2 (double copy) and loss less than 0.5 (one-half copy). Thirty-five copy gains and 38 copy losses were identified in this study. Copy gain was identified in 30 genes from tissue DNA, 1 gene from BALF EV DNA, and 2 genes from both tissue and BALF EV DNAs. Copy loss was exhibited in 19 genes in from tissue DNA, 15 genes from BALF EV DNA, and 7 from both tissue and BALF EV DNAs. The sensitivity of BALF EV DNA was 6% for gain and 27% for loss, using CNVs detection in tissue DNA as a reference. The concordances were 6% for copy gain and 17% for copy loss. Figure S5 shows a plot of representative cases of copy gain in both tumor tissue-derived and BALF EV-derived DNA. The log2 ratio of MYC was 2.3 for tissue DNA and 1.5 for BALF EV DNA, and that of EGFR was 3.7 for tissue DNA and 2.1 for BALF EV DNA. From this result, it can be inferred that the lower absolute value of BALF EV DNA is attributable to low tumor purity. Accurate estimation of CNVs in BALF EV DNA is challenging and is more difficult than identifying low AF mutations.
Discussion
In this study, we investigated the utility of BALF-EV as a reliable DNA source for targeted NGS analysis for the detection and quantification of mutations in 20 patients with EGFR-mutated lung adenocarcinoma and compared its performance with that of DNA obtained from matched FFPE tissue samples.
In terms of quality statistics, low DNA yield and low tumor purity were observed for BALF EV DNA compared with tissue DNA. Tumor purity and mutational clonality are known to influence sequencing coverage (23). Low tumor purity resulted in the reduction of effective coverage of variant alleles in tumor cells. It was technically difficult to obtain BALF EV DNA with high tumor purity in the present study because current limitation in technology makes obtaining tumor-specific EVs from various EVs present in the BALF samples. Further research is required to establish protocols for obtaining EV with high tumor purity. Increasing the amount of BALF used in NGS can yield higher BALF EV DNA concentration, thereby improving tumor purity and mean depth. In addition, we believe that the development of a method to selectively sort tumor-specific EVs from the BALF samples can also increase tumor purity, thereby achieving high effective coverage.
The DNA fragment length in BALF-EV was longer but the difference in library size was not statistically significant between those created using tissue and BALF EV DNAs. These results revealed that the integrity of BALF EV DNA was greater than that of tissue DNA. Larger library fragments could result in greater coverage variability across the target region (31). In this study, the molecular complexity of a genomic sequencing library was maintained in BALF EV DNA, and our findings demonstrate that BALF EV DNA has sufficient quality for use in NGS analysis.
In identifying a candidate mutation, the concordance rate between tissue and BALF EV DNAs was increased when variants with “benign” or “likely-benign” ClinVar clinical significance value was excluded. These findings demonstrated that targeted NGS using the BALF EV DNA has high clinical feasibility and utility.
We unfortunately identified 4 discordant cases, which were found to harbor exon 19 deletion in the tissue DNA NGS, but none in the corresponding BALF EV DNA NGS. It was difficult to distinguish between the true absence of the mutation in BALF EV DNA and technical error. To determine the lower limit-of-detection (LOD), the lowest concentration of an analyte that can be detected reliably (typically defined as having 95% sensitivity) of variants based on dilution assays was needed. The depth of coverage needed to maintain a given sensitivity, increases greatly as VAF decreases. Therefore, in the future studies we should examine the sensitivity for a somatic SNV of given VAF as a function of sequencing depth. These 4 cases showed similar or higher values for DNA quantity, tumor purity, and sequencing depth compared with the 11 exon 19del concordant cases. We extracted unmapped reads of these 4 cases, which were discarded before variant calling. We generated a new exon 19del reference sequence by removing deleted sequences and joining both ends. The extracted unmapped reads were re-mapped against new reference sequences, but we could not find any additional exon 19del mutations. Hasan et al. developed a tool, genesis-indel, which can find missed indels from unmapped reads (32). We used this tool, but obtained no additional indels. Thus, we are unable to explain the cause of these 4 discordant cases.
In NGS-based liquid biopsy, the ctDNA AF in the plasma is much lower than that in tumor tissues due to dilution of ctDNA within the cfDNA extracted from normal cells. In line with this notion, in the present study, the VAF in BALF EV DNA was much lower than (<5%) that in tissue DNA. Inadequate sequencing depth and low tumor content can contribute to false negative results in liquid NGS. Therefore, our findings highlight the need for a platform with high sensitivity for detection of low VAF in BALF EV DNA. Much higher sequencing coverage was needed due to low VAF.
In the present, the NGS results obtained using BALF EV DNA achieved concordance comparable to or higher than that of ctDNA with lower sequencing depth. An NGS study using ctDNA suggested ultra-high depth sequencing with median depth of 2,000× can detect low allele fraction variants (33). In our study, BALF EV DNA showed 81% concordance with tissue DNA, with median sequencing depth of 379x.
We also assessed the analytic performance of BALF EV DNA in determining the TMB, a biomarker to predict the response to immune checkpoint inhibitors. Results showed a positive correlation between the TMB scores in tissue and BALF EV DNAs, demonstrating the usefulness of BALF EV DNA in determination of TMB.
We obtained low concordance of CNV between tissue and BALF EV DNAs. Identification of CNV in targeted sequencing is challenging. In particular, for samples with low tumor purity, defining the limit values of copy loss or gain is more difficult, and numerous false positive or negative results are obtained in the process of distinguishing between the real copy variants and background noise.
Conclusions
In conclusion, to our knowledge, this is the first study to perform a comprehensive molecular profiling in a clinical NGS panel using EV DNA from BALF and corresponding tumor tissue biopsies from patients with lung adenocarcinoma. Although DNA yield from BALF-EV was low and needed much higher sequencing coverage and greater optimization of the NGS-pipeline to enable detection of low-frequency variants, the quality and quantity of BALF EV DNA were sufficient for NGS with comparable results to tissue DNA. It is logical to assume that tumor specific DNA in BALF EV DNA could be diluted by EVs released from other cells forming the TME, such as immune cells and alveolar epithelial cells etc. However, high concordance rate reflects that EV DNA have enough tumor specific DNA for NGS analysis. This study demonstrates that targeted NGS using BALF EV DNA for detecting actionable genetic alterations has high clinical feasibility and utility. In addition, further development of standardized technology that would allow easier access to isolation of EVs would be required to adapt this technique to clinical practices.
Acknowledgments
We thank Dr. Min Kyo Jung for the electron microscopy image of BALF-EVs.
Funding: The study was supported by the CJ Healthcare, Ltd. (CS2017_0029).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-888
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-888
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-888). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the amended Declaration of Helsinki (as revised in 2013). The study protocol was approved by the institutional review board of Konkuk University Medical Center (KUH1010899), and written informed consents were obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ettinger DS, Aisner DL, Wood DE, et al. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 5.2018. J Natl Compr Canc Netw 2018;16:807-21. [Crossref] [PubMed]
- Hagemann IS, Devarakonda S, Lockwood CM, et al. Clinical next-generation sequencing in patients with non-small cell lung cancer. Cancer 2015;121:631-9. [Crossref] [PubMed]
- Al-Kateb H, Nguyen TT, Steger-May K, et al. Identification of major factors associated with failed clinical molecular oncology testing performed by next generation sequencing (NGS). Mol Oncol 2015;9:1737-43. [Crossref] [PubMed]
- Kim HR, Lee SY, Hyun DS, et al. Detection of EGFR mutations in circulating free DNA by PNA-mediated PCR clamping. J Exp Clin Cancer Res 2013;32:50. [Crossref] [PubMed]
- Taniguchi K, Uchida J, Nishino K, et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res 2011;17:7808-15. [Crossref] [PubMed]
- Pender A, Garcia-Murillas I, Rana S, et al. Efficient Genotyping of KRAS Mutant Non-Small Cell Lung Cancer Using a Multiplexed Droplet Digital PCR Approach. PLoS One 2015;10:e0139074. [Crossref] [PubMed]
- Xu S, Lou F, Wu Y, et al. Circulating tumor DNA identified by targeted sequencing in advanced-stage non-small cell lung cancer patients. Cancer Lett 2016;370:324-31. [Crossref] [PubMed]
- Chen KZ, Lou F, Yang F, et al. Circulating Tumor DNA Detection in Early-Stage Non-Small Cell Lung Cancer Patients by Targeted Sequencing. Sci Rep 2016;6:31985. [Crossref] [PubMed]
- Mouliere F, Robert B, Arnau Peyrotte E, et al. High fragmentation characterizes tumour-derived circulating DNA. PLoS One 2011;6:e23418. [Crossref] [PubMed]
- Zhang YC, Zhou Q, Wu YL. The emerging roles of NGS-based liquid biopsy in non-small cell lung cancer. J Hematol Oncol 2017;10:167. [Crossref] [PubMed]
- Lee JS, Hur JY, Kim IA, et al. Liquid biopsy using the supernatant of a pleural effusion for EGFR genotyping in pulmonary adenocarcinoma patients: a comparison between cell-free DNA and extracellular vesicle-derived DNA. BMC Cancer 2018;18:1236. [Crossref] [PubMed]
- Hur JY, Kim HJ, Lee JS, et al. Extracellular vesicle-derived DNA for performing EGFR genotyping of NSCLC patients. Mol Cancer 2018;17:15. [Crossref] [PubMed]
- Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol 2002;2:569-79. [Crossref] [PubMed]
- Muralidharan-Chari V, Clancy JW, Sedgwick A, et al. Microvesicles: mediators of extracellular communication during cancer progression. J Cell Sci 2010;123:1603-11. [Crossref] [PubMed]
- Lazaro-Ibanez E, Lasser C, Shelke GV, et al. DNA analysis of low- and high-density fractions defines heterogeneous subpopulations of small extracellular vesicles based on their DNA cargo and topology. J Extracell Vesicles 2019;8:1656993. [Crossref] [PubMed]
- San Lucas FA, Allenson K, Bernard V, et al. Minimally invasive genomic and transcriptomic profiling of visceral cancers by next-generation sequencing of circulating exosomes. Ann Oncol 2016;27:635-41. [Crossref] [PubMed]
- Kahlert C, Melo SA, Protopopov A, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem 2014;289:3869-75. [Crossref] [PubMed]
- Thakur BK, Zhang H, Becker A, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res 2014;24:766-9. [Crossref] [PubMed]
- Allenson K, Castillo J, San Lucas FA, et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann Oncol 2017;28:741-7. [Crossref] [PubMed]
- Wan Y, Liu B, Lei H, et al. Nanoscale extracellular vesicle-derived DNA is superior to circulating cell-free DNA for mutation detection in early-stage non-small-cell lung cancer. Ann Oncol 2018;29:2379-83. [Crossref] [PubMed]
- Kim YT, Kim JW, Kim SK, et al. Simultaneous genotyping of multiple somatic mutations by using a clamping PNA and PNA detection probes. Chembiochem 2015;16:209-13. [Crossref] [PubMed]
- Han JY, Choi JJ, Kim JY, et al. PNA clamping-assisted fluorescence melting curve analysis for detecting EGFR and KRAS mutations in the circulating tumor DNA of patients with advanced non-small cell lung cancer. BMC Cancer 2016;16:627. [Crossref] [PubMed]
- Shin HT, Choi YL, Yun JW, et al. Prevalence and detection of low-allele-fraction variants in clinical cancer samples. Nat Commun 2017;8:1377. [Crossref] [PubMed]
- Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285-91. [Crossref] [PubMed]
- Fu W, O'Connor TD, Jun G, et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 2013;493:216-20. [Crossref] [PubMed]
- 1000 Genomes Project Consortium; Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012;491:56-65.
- Health KNIo. Korean Reference Genome. 2016. Available online: http://www.nih.go.kr/NIH/eng/contents/NihEngContentView.jsp?cid=65513&menuIds=HOME004-MNU2210-MNU2245-MNU2251-MNU2252
- Lee S, Seo J, Park J, et al. Korean Variant Archive (KOVA): a reference database of genetic variations in the Korean population. Sci Rep 2017;7:4287. [Crossref] [PubMed]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [Crossref] [PubMed]
- Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202-6. [Crossref] [PubMed]
- Spencer DH, Sehn JK, Abel HJ, et al. Comparison of clinical targeted next-generation sequence data from formalin-fixed and fresh-frozen tissue specimens. J Mol Diagn 2013;15:623-33. [Crossref] [PubMed]
- Hasan MS, Wu X, Zhang L. Uncovering missed indels by leveraging unmapped reads. Sci Rep 2019;9:11093. [Crossref] [PubMed]
- Malapelle U, Pisapia P, Rocco D, et al. Next generation sequencing techniques in liquid biopsy: focus on non-small cell lung cancer patients. Transl Lung Cancer Res 2016;5:505-10. [Crossref] [PubMed]


