
Prognostic and predictive effect of KRAS gene copy number and mutation status in early stage non-small cell lung cancer patients
Introduction
Advances in next generation sequencing have led to the identification of molecular alterations in non-small cell lung cancer (NSCLC), including mutations, fusions, increased tumor mutation burden, and copy number aberrations (CNA), among others. However, understanding the complex genetic landscape in NSCLC remains challenging, as only a small proportion of these alterations likely have clinical significance.
Previous LACE-Bio studies evaluated the prognostic and predictive role of KRAS mutations, and KRAS/TP53 co-mutations in early stage NSCLC (1,2). No clear prognostic or predictive role was detected, although a potentially negative predictive effect of TP53/KRAS co-mutation (2), and a possible deleterious effect of adjuvant chemotherapy in patients with a KRAS codon-13 mutation require further investigation (1). The KRAS oncogene is mutated frequently in NSCLC, particularly adenocarcinoma (3-5); however, the clinical impact of KRAS CNAs is unclear. Evaluation of the genome of primary lung adenocarcinomas by Weir et al. revealed areas of copy number amplification at 12p12.1, a region known to harbor the KRAS oncogene (6). Likewise, characterization of gene CNAs in resected NSCLC by the LACE-Bio consortium showed KRAS copy number gains; however, no significant association between KRAS CNA and survival was observed (7).
Few studies have shown that KRAS copy number gain might be associated with the presence of concomitant KRAS mutations (6,8,9). For instance, Modrek et al. showed that KRAS-mutated tumors appeared to have higher copy number compared to wild type (WT) tumors, with distinct patterns of gain across chromosome 12 (9). Moreover, even moderate KRAS copy number gains have been associated with increases in KRAS mRNA expression (8,9).
The prognostic and predictive role of concomitant KRAS mutations and CNA has not been well elucidated. Sasaki et al. showed that NSCLC patients with KRAS mutated tumors and increased copy number had worse prognosis compared to those with KRAS WT tumors with no increase in copy number; however, their study was limited by a small number of patients (10). In the current study, we completed a pooled analysis of LACE-Bio data to characterize the prognostic effect of KRAS mutation status and concomitant CN gain in early stage NSCLC, and to determine the ability to predict survival benefit from adjuvant chemotherapy. We hypothesized that concomitant KRAS mutations and CN gain would be prognostic of worse survival compared to KRAS mutations alone and that patients with mutation and CN gain might have the greatest potential to benefit from adjuvant chemotherapy.
We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-927).
Methods
Study design/patient characteristics
The LACE (Lung Adjuvant Cisplatin Evaluation)-Bio group conducts meta-analyses of data from 3 adjuvant randomized clinical trials [JBR.10, IALT (International Adjuvant Lung Cancer Trial), ANITA (Adjuvant Navelbine International Trialist Association)] comparing cisplatin-based chemotherapy versus observation, and CALGB-9633, which compared carboplatin-based chemotherapy versus observation (11-16). The LACE-Bio collaboration includes bio-banked tissue samples prospectively collected from JBR.10 and CALGB-9633, as well as retrospective samples from the IALT group (7,17). In this study, clinical and genomic data from the LACE-Bio consortium were used to evaluate the prognostic and predictive effect of concomitant KRAS mutation and gene CNAs in early stage NSCLC patients. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013).
KRAS gene copy number and mutation analyses
DNA extracted from formalin-fixed and paraffin-embedded (FFPE) sections was used for KRAS mutation and genome-wide copy number analyses using methods previously described (1,7). Briefly, KRAS mutation analyses were completed using allelic specific oligonucleotide hybridization with sequencing confirmation in JBR.10, and PCR amplification and direct sequencing of exon 2 in IALT and CALGB-9633 (1). Whole-genome copy number analyses were completed using the OncoScan CNV Plus Assay (ThermoFisher, Carlsbad, California, USA) (7). CNAs were categorized into two groups: Gain (2-fold higher CN) or Neutral (Neut)/Loss; and KRAS mutation status was defined as WT or mutant (MUT). The following groups were compared in all patients and the adenocarcinoma subgroup: WT + Neut/Loss (reference), WT + Gain, MUT + Gain, MUT + Neut/Loss.
Outcomes
The primary endpoint was lung cancer-specific survival (LCSS), defined as the time from randomization to death from lung cancer, censoring at the date of death from other causes and without lung cancer recurrence. Secondary endpoints included disease free survival (DFS), defined as the time from randomization to the time of first event (recurrence or death from any cause); and overall survival (OS), defined as the time from randomization to the date of death from any cause. Patients for whom no events had been observed were censored at the date of their last follow-up.
Statistical analysis
The Kaplan-Meier method and log-rank test were used to compare survival curves between the four groups mentioned above. KRAS mutation status, and concomitant KRAS CNA and mutational status were separately correlated to survival endpoints using a Cox proportional hazards model, which was stratified by trial in all models, adjusted for treatment in the unadjusted model, and adjusted for treatment, age, sex, tumor stage, nodal stage, histology, WHO performance status and surgery type in the fully adjusted model. For KRAS mutation status, the WT group was the reference category, and the HR was reported for MUT versus WT group. For the concomitant KRAS CNA and mutation status, WT + Neut/Loss was the reference category, and the HR for a given category (MUT + Gain, MUT + Neut/Loss or WT + Gain) was interpreted as the relative hazard for that category versus the WT + Neut/Loss category. To evaluate the predictive role of concomitant KRAS CNA and mutation status, a treatment-by-variable interaction was added to the Cox models stratified by study. For each endpoint, we compared the treatment effect across four concomitant KRAS CNA and mutation status groups by using the ratio of HRs (rHR): the ratio of the HR for adjuvant chemotherapy versus observation in a given category ‘c’ (MUT + Gain, MUT + Neut/Loss or WT + Gain) to the HR for adjuvant chemotherapy versus observation in the WT + Neut/Loss category (rHR = HR(Chemo vs. Obs. in c)/HR(Chemo vs. Obs. in WT + Neut/Loss)). A rHR <1.0 indicates that the treatment effect size was greater for category ‘c’ than for WT + Neut/Loss patients. Given that KRAS mutations occur primarily in the adenocarcinoma group, analyses were also performed separately for this subtype. Statistical significance was set at P<0.05. All statistical analyses were performed using the ‘Survival’ and the ‘survminer’ packages in R software version 3.6.2.
Results
Patient characteristics
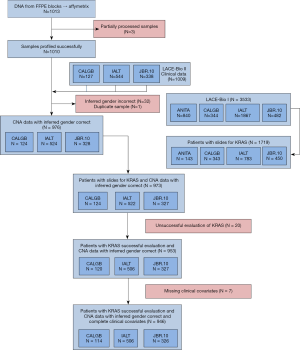
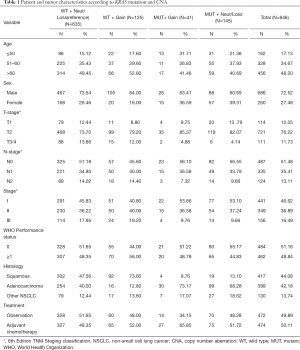
A total of 946 patients had complete clinical data including both KRAS mutation and CNA results (Figure 1): 41 MUT + Gain, 145 MUT + Neut/Loss, 125 WT + Gain, and 635 WT + Neut/Loss (reference group). There were 399 patients in the adenocarcinoma subgroup, with 30 MUT + Gain, 99 MUT + Neut/Loss, 16 WT + Gain, and 254 WT + Neut/Loss. The most common KRAS mutation subtypes were KRAS G12C (n=89; 47.8%), G12V (n=44; 23.6%) and G12D (n=18; 9.7%) (Table S1). Baseline patient characteristics for the total population are summarized in Table 1. Almost half of the patients were older than 60 years of age, with the majority (72.52%) being male. Data regarding smoking history were collected in the JBR.10 and CALGB-9633 trials, and summarized in Supplementary Table 2. Of these 440 patients, 397 (90.2%) were current/former smokers. Moreover, all KRAS MUT + Gain patients (n=25) were current/formers smokers (Tables S2,S3). Staging included 46.62% with stage I, 36.89% stage II and 16.49% stage III. There was a similar proportion of patients with adenocarcinoma (42.18%) and squamous cell carcinoma (44.08%). Adjuvant chemotherapy was given to half of patients. There were no significant differences in baseline patient or tumor characteristics among the four subgroups.
Full table
Full table
Prognostic effect
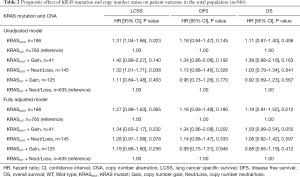
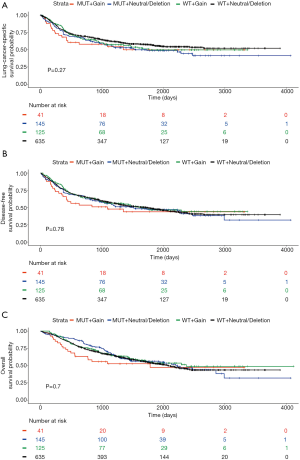
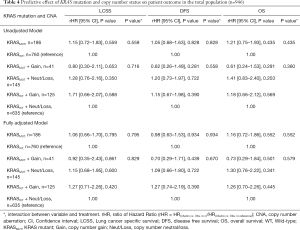
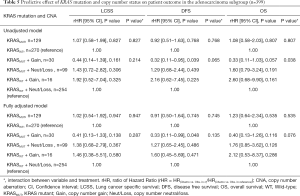
Kaplan-Meier analyses showed no significant differences in terms of LCSS, DFS or OS among the four groups based on KRAS CNAs and mutational status in the total population (Figure 2), or in the adenocarcinoma subgroup (data not shown). Univariable analyses showed that KRAS MUT and KRAS MUT + Neut/Loss were associated with poor LCSS outcome: the hazard ratio for patients with a KRAS MUT was 1.31 (95% CI, 1.04–1.66, P=0.023) relative to the KRAS WT group, and the hazard ratio for patients with KRAS MUT + Neut/Loss was 1.32 (95% CI, 1.01–1.71, P=0.038) relative to the KRAS WT + Neut/Loss category (Table 2). However, this was no longer significant after adjusting for clinical covariates (KRAS MUT HR =1.27; 95% CI, 0.98–1.63, P=0.065; and KRAS MUT + Neut/Loss HR =1.28; 95% CI, 0.97–1.68, P=0.078, respectively) (Table 2). In both univariable and multivariable analyses including all patients, the worst outcomes consistently were observed in the KRAS MUT + Gain subgroup for LCSS (HR =1.34; 95% CI, 0.83–2.17, P=0.23), DFS (HR =1.34; 95% CI, 0.86–2.09, P=0.20,) and OS (HR =1.59; 95% CI, 0.99–2.54, P=0.055), compared to WT + Neut/Loss (Table 2). When limiting analyses to KRAS MUT + Gain compared directly to KRAS WT + Neut/Loss, there was a significant negative prognostic effect of KRAS MUT + Gain for OS (HR =1.66; 95% CI, 1.03–2.67, P=0.037; Table 3).

Full table
In the adenocarcinoma subgroup, there was no significant prognostic effect of KRAS mutation and CNA on LCSS, DFS or OS (Table S4). However, there was a non-significant trend towards worse OS (HR =1.51; 95% CI, 0.84–2.72, P=0.169) when KRAS MUT + Gain was compared directly to KRAS WT + Neut/Loss in adenocarcinoma patients (results not shown).
Predictive effect
Results of the univariable and multivariable analysis did not show any significant predictive effect of KRAS CNA and mutation status in the overall population (Figure 3A,B,C and Table 4) (interaction P=0.829, 0.670 and 0.579 for LCSS, DFS and OS, respectively, in multivariable analysis). In the adenocarcinoma subgroup, univariable analysis showed a significant predictive effect of KRAS CNA and mutational status for OS (interaction P=0.038), although this was no longer significant after adjusting for clinical covariates (interaction P=0.076; Table 5). However, there appeared to be a beneficial effect of chemotherapy on DFS and OS based on Kaplan-Meier analysis in KRAS MUT + Gain adenocarcinoma patients when compared to observation (Figure 3D,E,F).
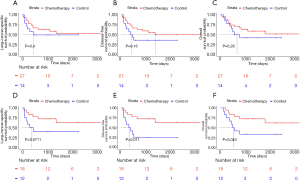
Full table
In addition, univariable Cox regression models in the adenocarcinoma population showed that the beneficial effect of chemotherapy on DFS was significantly more pronounced for the KRAS MUT + Gain group compared to WT + Neut/Loss patients (rHR =0.32; 95% CI, 0.11–0.94, P=0.039), with non-significant trends for better LCSS (rHR =0.44; 95% CI, 0.14–1.39, P=0.161) and OS (rHR =0.33; 95% CI, 0.11–1.03, P=0.057). Similarly, multivariable analysis showed a significantly higher treatment effect size in KRAS MUT + Gain patients for DFS (rHR =0.33; 95% CI, 0.11–0.99, P=0.048) in adenocarcinoma patients (Table 5), with a non-significant trend also seen for LCSS (rHR =0.41; 95% CI, 0.13–1.33, P=0.138) and OS (rHR =0.40; 95% CI, 0.13– 1.26, P=0.116). Similar trends were also noted in the total population (Table 4), with a suggestion of more benefit to improve DFS (rHR =0.70; 95% CI, 0.29–1.71, P=0.439) and OS (rHR =0.73; 95% CI, 0.29–1.84, P=0.501) with adjuvant chemotherapy in the KRAS MUT + Gain group relative to WT + Neut/Loss patients; this higher benefit was likely driven by the adenocarcinoma subset.
Full table
Discussion
The KRAS oncogene frequently is mutated in NSCLC, particularly adenocarcinoma, and the biologic heterogeneity of KRAS-mutated lung cancer has been well characterized. The majority of KRAS mutations occur at codon 12 (18), and different KRAS point mutations are associated with diversity in biological behavior, response to therapy, and clinical outcome (5). However, the role of gene CNAs in KRAS-mutated tumors is not as well characterized. A correlation between KRAS mutations and copy number gain has been reported in NSCLC (6,8,9); however, these studies have been limited by small numbers of patients. A study by Modrek et al. found concomitant KRAS mutations and copy number gain in 11 out of 166 (6.6%) NSCLC patients (9). Likewise, Sasaki et al. evaluated 172 surgical tissue samples from NSCLC patients and identified 8 patients (4.6%) with concomitant KRAS mutation and gene copy number gain (10). In our pooled analysis of 946 patients, we were able to identify 41 patients (4.3%) in the total population who had a KRAS mutation with concomitant copy number gain, which is consistent with previous reports. Overall, the proportion of patients who harbor concomitant KRAS mutation and copy number gain is quite small (approximately 4–6%), suggesting that this is an uncommon occurrence.
To our knowledge, this study is the largest to report on the prognostic and predictive roles of concomitant KRAS mutation and copy number gain. We observed a consistent but non-significant trend towards worse LCSS (HR =1.34; 95% CI, 0.83–2.17, P=0.232), DFS (HR =1.34; 95% CI, 0.86–2.09, P=0.202) and OS (HR =1.59; 95% CI, 0.99–2.54, P=0.055) in KRAS MUT + Gain patients relative to the WT + Neut/Loss group in the total population. The adenocarcinoma subgroup accounted for 73.2% (n=30) of the MUT + Gain patients, and although there was no significant prognostic effect noted, there appeared to be a similar trend towards worse OS (HR =1.40; 95% CI, 0.78–2.51, P=0.255). Sasaki et al. previously reported worse prognosis in NSCLC patients with both KRAS mutations and increased gene copy number when compared to KRAS WT patients with no increase in copy number; however, the study was limited by a small number of patients (n=8) (10). When we completed a similar comparison limited to only the two subgroups of KRAS MUT + Gain compared to WT + Neut/Loss, we also observed a significantly worse OS (HR =1.66; 95% CI, 1.02–2.67, P=0.037) in KRAS MUT + Gain patients.
Singh et al. showed a significant correlation between KRAS gene amplification and KRAS-dependency. Moreover, increased gene copy number in KRAS-mutated tumors has been associated with elevated KRAS mRNA and protein expression (9,19). Perhaps there is a selection for copy number gain in tumors that already harbor a KRAS mutation, thereby conferring dependency on the KRAS oncogene for growth and survival. Studies suggest that KRAS oncogene addiction might play a role in early tumor initiation, with preclinical data showing development of early onset lung cancer in mice harboring mutations in KRAS (20). However, although KRAS mutations alone have been shown to promote lung tumor formation, additional genomic alterations are likely required for ongoing tumor progression (21). It is plausible that gene amplification is acquired in KRAS-dependent tumors to aid in tumor progression. Newnham et al. compared resected and recurrent NSCLC tumors and showed differential gene expression suggesting that concomitant KRAS mutations with CN gain might be implicated in tumor recurrence clinically (3). Moreover, preclinical models show that copy number gain in KRAS-mutated tumors appears to select for enhanced glycolytic metabolism (a compensatory mechanism to promote survival in tumors with mitochondrial defects), and might also be involved in increasing metastatic potential in lung cancer cells (21). Interestingly, we previously showed an increased risk (HR =2.76; 95% CI, 1.34–5.70, P=0.005) of developing a second primary cancer in patients with KRAS-mutated tumors (1). The impact of concomitant KRAS mutation and gene copy number gain on cell survival, tumor recurrence, or the development of second primary cancers is unclear, and should be further investigated to determine if this might explain the observed trend towards worse survival in KRAS MUT + Gain patients.
Variable responses to chemotherapy have been reported in KRAS-mutated tumors, and may depend on the subtype of the KRAS mutation (22,23). Garassino et al. reported a decreased response to cisplatin but increased response to paclitaxel and pemetrexed in KRAS G12C mutated tumors, whereas, KRAS G12D mutations appeared to be resistant to paclitaxel, and G12V mutations were highly sensitive to cisplatin but slightly more resistant to pemetrexed (23). Shepherd et al. previously reported variability in the effect of adjuvant chemotherapy on survival based on KRAS mutation subtypes, with worse DFS (HR =3.88; 95% CI, 1.44–10.46, P=0.007, interaction P=0.01) and OS (HR =5.78; 95% CI, 2.06–16.2, P=0.001, interaction P=0.002) observed in patients with KRAS codon 13 mutations who received adjuvant chemotherapy (1). In addition, a potential deleterious effect of adjuvant chemotherapy was seen in patients with KRAS/TP53 co-mutations when compared to WT/WT tumors (HR =2.49; 95% CI, 1.10–5.64, P=0.03) (2). However, due to small patient numbers, these data require further validation. In contrast, we found that KRAS MUT + Gain was associated with a higher beneficial effect of chemotherapy on DFS (rHR =0.33; 95% CI, 0.11–0.99, P=0.048) compared to the WT + Neut/Loss group in adenocarcinoma patients, with a non-significant trend also seen for LCSS (rHR =0.41; 95% CI, 0.13–1.33, P=0.14) and OS (rHR =0.40; 95% CI, 0.13–1.26, P=0.12) on multivariable analysis. A similar higher beneficial effect of chemotherapy on DFS and OS was also noted in KRAS MUT + Gain patients compared to WT + Neut/Loss patients in the total population, but this benefit may be driven by the adenocarcinoma subset. It is possible that KRAS MUT + Gain patients have an increased dependency on KRAS activation, which might lead to increased susceptibility to chemotherapy, leading to a decrease in risk for tumor recurrence. Whether this hypothesis might explain the higher beneficial effect of chemotherapy on DFS observed in our study is still to be determined, and additional studies are required to elucidate the mechanism by which KRAS MUT-Gain patients might be more responsive to chemotherapy.
There are limitations in this study. The KRAS mutation analyses were pooled from separate trials; therefore, there were differences in technique and laboratories used for testing. Secondly, we were unable to evaluate the predictive or prognostic role of different KRAS mutation subtypes in patients with KRAS mutations and CNA due to the small numbers in each group. This further highlights the need for pooled analyses to ensure adequate sample numbers for robust analyses for less common genetic alterations. Finally, we did not have complete smoking history data from all trials included in the current study; therefore, survival analyses adjusting for smoking status could not be completed due to the small sample size.
Conclusions
In summary, the current LACE-Bio analysis showed that concomitant KRAS mutations and copy number gain are rare, occurring in approximately 4% of NSCLC patients. Although not significant, there was a trend towards worse LCSS, DFS and OS in KRAS MUT + Gain patients relative to WT + Neut/Loss patients. In addition, concomitant KRAS mutation and CN gain appeared to predict for better DFS in adenocarcinoma patients treated with adjuvant chemotherapy, with a trend towards better LCSS and OS as well. However, these results could be driven by the small number of patients and require further validation.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-927
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-927). Elisabeth Brambilla and Dr Ming-Sound Tsao serves as an unpaid editorial board member of Translational Lung Cancer Research. LS reports a GSK grant to institution; JCS reports personal fees from Abbvie, AstraZeneca, Bayer, Blend Therapeutics, Boeringher Ingelheim, Cytomix, Daiichi Sankyo, Eli Lilly, Genmab, Guardiant Health, Inivata, Merck, Netcancer, Roche, Servier, Pharmamar, Tarveda outside the submitted work, and was a full time employee of AstraZeneca from Sept 2017 to December 2019; RG reports personal fees from Millennium Pharmaceuticals, AbbVie, F. Hoffmann La-Roche, EMD Sereno, Genenetech, Janssen, Bristol Meyers Squibb, Eli Lilly, AstraZeneca, Pfizer, Celgene, Partner Therapeutics, GSK, Merck, Jounce, Amgen, Achilles, Horizon Pharmaceuticals, GenePlus, outside the submitted work; SM reports personal fees from IDDI Belgium, Hexal, Steba, IQVIA, Roche, Sensorion, Biophytis, Servier, Yuhan, outside the submitted work. The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Shepherd FA, Domerg C, Hainaut P, et al. Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J Clin Oncol 2013;31:2173-81. [Crossref] [PubMed]
- Shepherd FA, Lacas B, Le Teuff G, et al. Pooled Analysis of the Prognostic and Predictive Effects of TP53 Comutation Status Combined With KRAS or EGFR Mutation in Early-Stage Resected Non-Small-Cell Lung Cancer in Four Trials of Adjuvant Chemotherapy. J Clin Oncol 2017;35:2018-27. [Crossref] [PubMed]
- Newnham GM, Conron M, McLachlan S, et al. Integrated mutation, copy number and expression profiling in resectable non-small cell lung cancer. BMC Cancer 2011;11:93. [Crossref] [PubMed]
- Suh JH, Johnson A, Albacker L, et al. Comprehensive Genomic Profiling Facilitates Implementation of the National Comprehensive Cancer Network Guidelines for Lung Cancer Biomarker Testing and Identifies Patients Who May Benefit From Enrollment in Mechanism-Driven Clinical Trials. Oncologist 2016;21:684-91. [Crossref] [PubMed]
- Yang H, Liang SQ, Schmid RA, et al. New Horizons in KRAS-Mutant Lung Cancer: Dawn After Darkness. Front Oncol 2019;9:953. [Crossref] [PubMed]
- Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature 2007;450:893-8. [Crossref] [PubMed]
- Rotolo F, Zhu CQ, Brambilla E, et al. Genome-wide copy number analyses of samples from LACE-Bio project identify novel prognostic and predictive markers in early stage non-small cell lung cancer. Transl Lung Cancer Res. 2018;7:416-27. [Crossref] [PubMed]
- Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008;455:1069-75. [Crossref] [PubMed]
- Modrek B, Ge L, Pandita A, et al. Oncogenic activating mutations are associated with local copy gain. Mol Cancer Res 2009;7:1244-52. [Crossref] [PubMed]
- Sasaki H, Hikosaka Y, Kawano O, et al. Evaluation of Kras gene mutation and copy number gain in non-small cell lung cancer. J Thorac Oncol 2011;6:15-20. [Crossref] [PubMed]
- Winton T, Livingston R, Johnson D, et al. Vinorelbine plus cisplatin vs. Observation in resected non-small-cell lung cancer. N Engl J Med 2005;352:2589-97. [Crossref] [PubMed]
- Butts CA, Ding K, Seymour L, et al. Randomized phase III trial of vinorelbine plus cisplatin compared with observation in completely resected stage IB and II non-small cell lung cancer: Updated survival analysis of JBR-10. J Clin Oncol 2010;28:29-34. [Crossref] [PubMed]
- Arriagada R, Bergman B, Dunant A, et al. Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N Engl J Med 2004;350:351-60. [Crossref] [PubMed]
- Arriagada R, Dunant A, Pignon JP, et al. Long-term results of the international adjuvant lung trial evaluating adjuvant cisplatin-based adjuvant chemotherapy in resected lung cancer. J Clin Oncol 2010;28:35-42. [Crossref] [PubMed]
- Douillard JY, Rosell R, De Lena M, et al. Adjuvant vinorelbine plus cisplatin versus observation in patients with completely resected stage IB-IIIA non-small cell lung cancer (Adjuvant Navelbine International Trialist Association [ANITA]): A randomised controlled trial. Lancet Oncol 2006;7:719-27. [Crossref] [PubMed]
- Pignon JP, Tribodet H, Scagliotti GV, et al. Lung adjuvant cisplatin evaluation: A pooled analysis by the LACE Collaborative Group. J Clin Oncol 2008;26:3552-9. [Crossref] [PubMed]
- Seymour L, Le Teuff G, Brambilla E, et al. LACE-Bio: Validation of predictive and/or prognostic immunohistochemistry/histochemistry-based biomarkers in resected non-small cell lung cancer. Clin Lung Cancer 2019;20:66-73.e6. [Crossref] [PubMed]
- Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res 2012;72:2457-67. [Crossref] [PubMed]
- Singh A, Greninger P, Rhodes D, et al. A gene expression signature associated with "K-Ras addiction" reveals regulators of EMT and tumor cell survival. Cancer Cell 2009;15:489-500. [Crossref] [PubMed]
- Johnson L, Mercer K, Greenbaum D, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 2001;410:1111-6. [Crossref] [PubMed]
- Kerr EM, Gaude E, Turrell FK, et al. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature 2016;531:110-3. [Crossref] [PubMed]
- Rodenhuis S, Boerrigter L, Top B, et al. Mutational activation of the K-ras oncogene and the effect of chemotherapy in advanced adenocarcinoma of the lung: a prospective study. J Clin Oncol 1997;15:285-91. [Crossref] [PubMed]
- Garassino MC, Marabese M, Rusconi P, et al. Different types of K-Ras mutations could affect drug sensitivity and tumor behaviour in non-small-cell lung cancer. Ann Oncol 2011;22:235-7. [Crossref] [PubMed]


