
Radiomics and artificial intelligence in lung cancer screening
Introduction
According to World Health Organization data, lung cancer is the deadliest of all known tumors. In 2018, total lung cancer cases reached 2.09 million, with 1.76 million related deaths reported (1). Such a high mortality rate can be reduced by early diagnosis and treatment. Screening programs have been shown to significantly increase the number of cases diagnosed at an early stage and have become highly recommended (2-5). During screening, a variety of medical imaging modalities can be applied; however, for the screening of lung nodules, computed tomography (CT) is the keystone imaging technique, while other available methods are of lesser importance. To guarantee the effectiveness of a screening program, each data set should be carefully analyzed. However, the demands of such in-depth analysis on the time of experienced staff is a problem. This factor is especially relevant to medical image analysis, in which experienced radiologists with a limited amount of time must analyze a large number of images for a single patient.
The task of radiologists is to identify all pulmonary nodules and examine their border, shape, location, and size, as well as to judge their type (solid, part-solid, or non-solid) (6-9). During lung cancer screening, in which there could be multiple nodules potentially measuring only a few millimeters in size, such a procedure is extremely challenging and time-consuming. One possible solution for this problem is to use artificial intelligence.
Artificial Intelligence (AI) refers to the use of a computer to simulate intelligent behavior with or without minor human intervention (10). AI is employed in many areas of medicine, including medical diagnosis, medical statistics, robotics, and human biology. In the case of lung cancer screening, a branch of artificial intelligence, namely machine learning, provides algorithms as an aid for radiologists. Such techniques could serve as a computer-aided diagnostic system for identifying candidate nodules and retrieving as much diagnostically relevant information as possible. This paper focuses on a comprehensive review of such algorithms, beginning with the simplest solutions developed, with a major emphasis on the current state-of-the-art in radiomics and deep learning algorithms.
Computer-aided detection systems for detection and diagnosis of pulmonary nodules
The algorithms used in the pulmonary nodule identification process are referred to under the common name of computer-aided detection systems, and are based on the following main steps: lung segmentation, and pulmonary nodule detection and classification.
The first step, lung segmentation, is performed by removing the background and unwanted areas from the input CT image to narrow the image region for further examination. Over the years, a number of algorithms have been developed for this purpose. The first approaches focused on two-dimensional (2D) (11) and three-dimensional (3D) (12,13) region growing algorithms. Other widely used algorithms are based on Lai et al.’s active contour model (14). Recently, deep learning algorithms have overtaken the classical approaches as being less sensitive and more accurate . The current state-of-the-art methods utilize statistical finite element analysis (15), or three-dimensional lung segmentation, improved by the adversarial neural network training, which was successfully implemented by Siemens Healthcare in their AI-RAD Companion framework (16). A summary of state-of-the-art algorithms for lung segmentation is provided in Table 1.

Full table
Nodule candidate detection, the second step in CT scan analysis, is performed to identify structures within the lung that are suspected of being malignant lung nodules. CT scans with examples of benign and malignant nodules are shown in Figure 1. A number of algorithms have been published to accomplish the nodule detection task, among which, multiple grey-level thresholding (22) is considered to be the best. However, algorithms based on shape (23) and template matching (24), as well as morphological approaches with convexity models (25) and filtering-based methods (26), are also capable of successfully detecting candidate nodules with high accuracy. In 2019, a polygonal approximation algorithm (27) was proposed, followed by a neuro-evolutionary schemeJeny (28) in 2020. Since 2016, deep learning networks have played an important role in nodule detection. These systems are discussed in dedicated paragraphs later in this paper. A review of algorithms for the detection of candidate lung nodules is presented in Table 2.


Full table
The application of each lung nodule detection algorithm mentioned in Table 2 results in a number of false-positive candidates, the lowest rate for which was reported by Cascio et al. in (30) (2.5 FP/scan), with Ozekes reporting the highest rate in (29) (13.38 FP/scan). The reduction of false positives is achieved by applying nodule feature extraction or nodule candidate classification approaches. Several new methods for feature extraction and nodule candidate classification have been published recently. Table 3 summarizes such algorithms together with their reported accuracy.

Full table
Despite all of the research conducted so far, there is still a great need to improve existing CAD algorithms for lung cancer diagnosis.
Radiomics in lung cancer diagnosis and therapy
In a previous paragraph, we reviewed existing CAD algorithms, pointing out their main drawback, which is a lack of strict definition of the set of features which can be used to determine whether an identified nodule is cancerous or benign. In parallel to the development of CAD systems, Lambin et al. (44) defined a new concept called radiomics. Radiomics is based on the extraction of a large number of features from a single image using data-characterization algorithms. Such features assist in identifying cancer characteristics hidden from the naked eye of a human expert. However, the radiomics image processing pipeline consists of more steps than simply feature extraction. The step taken prior to radiomic feature calculation is segmentation of the region of interest (ROI), which in most cases is performed manually due to the lack of an accurate “gold standard” technique for pulmonary nodule segmentation. The third part of each radiomics analysis is pulmonary nodule classification—the process of model selection to perform one of the following tasks: (I) categorization of the analyzed nodule into one of two groups: malignant or benign; (II) prediction of the response to therapy (primarily radiotherapy); or (III) prediction of the overall survival of the patient.
The standard flow of each radiomic analysis is shown in Figure 2.
Lambin and Kumar’s initial papers (45) utilized only a limited number of image-derived features, whereas Aerst’s 2014 study (46) proposed a well-defined set of features that are used in almost all radiomics applications. In general, radiomic features are grouped into the size and shape-based features (47,48); descriptors of the image intensity histogram (49,50); descriptors of the relationships between voxels (51); derived textures (52,53); textures extracted from filtered images (50,54); and fractal features (55).
The definitions of the abovementioned features, along with brief guidance on how to calculate their values, have also been described in the works of Galloway (56), Pentland (57), Amadsun (58), and Thibault (59).
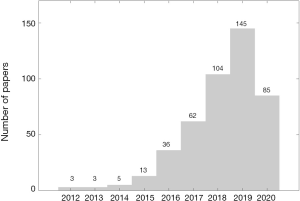
Over the years, radiomics has become one of the most popular and important techniques in the detection of a large variety of tumors. The increasing popularity of radiomics is clearly evidenced by the large number of papers containing the term “radiomics” together with “lung cancer” on the PubMed database. The numbers of papers available on PubMed from 2012 until May 2020 are presented in Figure 3.
The final step of model development is validation, in which the trained model is evaluated on new, independent data in order to check its performance. If the model achieves a reasonable performance on the validation data and performs as well as on the training data, its robustness and generalization are confirmed . Assuming representativeness of the training data set, a lower prediction performance on validation data would indicate overfitting (i.e., when a model draws false conclusions on training data that do not apply to new observations). A poor performance on both the training and validation data would indicate underfitting (i.e., when the classification model is unable to draw meaningful conclusions from the data).
Despite the potential of radiomics, as proven by the exponentially growing number of publications, the challenge of developing a general, robust signature that can be effectively implemented in a clinical setting still exists. This is reflected in the number of publications devoted to the problem surrounding the lack of reproducible radiomic signatures (60-62). Additionally, a number of authors have utilized test-retest procedures. As a result of such an approach, Zhovanic et al. (63) have shown that more than 60 radiomic features are sensitive to the different CT reconstruction parameters as well as to the specific vendors of different CT scanners. The research of Yip et al. (64) evidenced the relationship between the segmentation and stability of radiomic features, and it was subsequently concluded that radiomic features are sensitive to interobserver variability that exist naturally in the medical world. To deal with this problem, a set of guidelines called the “radiomics quality score” was proposed, including a 16-point checklist (consisting of robust segmentation, the stability of test-retest, description of the imaging protocol used, and internal and external validation, among other items) that should be submitted together with a radiomics study (65).
Moreover, Parmar et al. (66) showed that the choice of classification model for lung cancer assessment contributed the most to the variation in performance (34.21% of total variance). In their study, 12 different machine learning classifiers stemming from 12 classifier families (bagging, Bayesian, boosting, decision trees, discriminant analysis, generalized linear models, multiple adaptive regression splines, nearest neighbors, neural networks, partial least squares and principle component regression, random forests, and support vector machines) were tested on radiomic feature data. The researchers identified the random forest method to be the best for handling radiomic feature instability, achieving the highest prognostic performance.
In another study, Ferreira Junior et al. (67) used three different classification methods for the prediction of lung cancer histopathology and metastases. They used up to 100 radiomic features and evaluated the performance of the naïve Bayes method, the k-nearest neighbors algorithm, and a radial basis function-based artificial network. Although these methods are not widely used now, all showed great potential for the assessment of lung cancer by radiomics.
More recently, the success of artificial neural networks enabled the application of an end-to-end machine learning algorithm, which automatically extracts features from its input data. The development of convolutional neural networks (CNN), which are mathematical models devoted to imaging data, has shown great potential for their use in medical imaging. However, Tajbaksh et al. (68) showed that older neural network architectures outperformed 2D CNNs. The performance of the deep models was comparable to that of the shallow models, which were trained on previously extracted radiomic features.
Hosny et al. (69) extended the idea of applying CNNs from 2D to 3D data and demonstrated the potential of using deep learning for mortality risk stratification based on CT images from patients with non-small cell lung cancer (NSCLC).
Radiomics in lung cancer diagnosis
Radiomic features, calculated based on the low-dose computed tomography (LDCT) images, are frequently used in the screening and diagnosis of lung cancer. Multiple studies have shown that this approach supports the detection of lung cancer at an early stage, unlike molecular or blood tests. National Lung Screening Trial proved the effectiveness of radiomics in the early detection of malignant lung nodules, with the number of lung cancer-related deaths in the screening group 20% lower than that in the control group (70). Kumar et al. (71) demonstrated radiomics to be an effective tool in differentiating between malignant and benign tumors, with an accuracy of 79.06%, a sensitivity of 78.00%, and a specificity of 76.11% obtained from the Lung Image Database Consortium and Image Database Resource Initiative (LIDC-IRDI) dataset. Liu et al. (72) published their findings of four feature signatures that were able to differentiate between malignant and benign nodules with an accuracy of 81%, a sensitivity of 76.2%, and a specificity of 91.7%. In another study, conducted by Wu et al. (73), a 53-feature radiomic signature allowing for the classification of malignant and benign nodules with an area under the curve (AUC) equal to 72% was given.
The malignancy of a nodule is not the only important factor in the process of diagnosis and planning therapy—tumor stage is another. In the classical clinical approach, the tumor stage is estimated on the basis of a histopathological biopsy and other clinical factors. Chaddad et al. (74) identified radiomic features that are associated with the tumor, node, metastasis (TNM) stage of lung cancer patients. Wu et al. (75) proved that a radiomic-based feature allows for the identification of early-stage metastasis in lung cancer (M staging). Their findings were confirmed by Coroller et al. (76) in a study performed of 182 cases of lung adenocarcinoma.
Radiomics as a tool for predicting therapeutic response
The radiomic signature has proven to be effective not only in lung cancer detection and staging but also in predicting the response of the patient to the applied therapy. Aerts et al. (77) have shown that a radiomic signature evaluated before treatment aids in the prediction of EGFR-mutation related response to therapy among patients with NSCLC. Based on these findings, Coroller et al. (78) found that a radiomic signature could be used to predict response to chemoradiotherapy in NSCLC patients. Another successful application of radiomics was reported by Mattonen et al. (79), who predicted the risk of tumor recurrence based on analysis of radiomic features. Bogowicz et al. (80) stated that CT radiomics is promising pre-treatment and intra-treatment biomarker for therapeutic outcome prediction, although most of the published studies have been performed only in a retrospective setting. Recently, Vaidya et al. (81) published important results confirming that radiomics may be a promising tool for estimating the patient score to identify those who could benefit the most from adjuvant therapy. However, their most important finding was a correlation of radiomic features with multimodal biological data, which corroborates the relationship between radiomic features and tumor biology.
Despite the importance and success of radiomics in the fields discussed in the previous sections, the development of the alternative deep learning based methods performing feature extraction similar to radiomics is still a main research focus for many groups. Avanzo et al. (82) pointed out that deep learning can facilitate automated radiomic feature extraction without the need to design a set of hand-crafted radiomic features. However, they also noted that the explainability of deep learning models should be taken into account during model development, and further research should be conducted in that area.
Deep learning in lung cancer imaging
Deep neural networks are successfully used in many applications related to automated image recognition. In the analysis of lung cancer images, deep neural networks are primarily used to perform two key tasks: (I) detection and segmentation of pulmonary nodules; and (II) classification of identified pulmonary nodules.
Figure 4 presents a schematic diagram of how deep neural networks are being used in lung nodule detection (segmentation) (A) and classification (diagnosis) (B).
Deep learning-based detection and segmentation of pulmonary nodules
This section discusses the application of deep neural networks for the detection and segmentation of pulmonary nodules suspected to be malignant.
Khosravan et al. (83) designed a 3D CNN named S4ND (after Single-Shot Single-Scale lung Nodule Detection) to detect lung nodules without further processing. The input CT volume is divided into a 16×16×8 voxels grid, and each cell is passed through a 3D CNN comprising five dense blocks. The output is represented by a probability map of the presence of a nodule in each cell. LUNA16 was used as training data with 10-fold cross validation. On average, seven false-positive findings were produced per scan.
Golan et al. (84) used dense blocks only at the last layer of the convolution layer. As before, CT volume was divided into small sub-volumes of 5×20×20 voxels size. As whole or partial change of the nodule may be preserved in a cell, the network reads each sub-volume and interprets it in terms of nodule presence. An LIDC-IDRI dataset was used for training and cross-validation. The method produced an average of 20 false positives per scan. Zhu et al. (85) designed Deep 3D Dual Path Nets (3D DPN26) with 3D Faster R-CNN for nodule detection with 3D dual paths and a U-net-like structure for feature extraction. Dual paths are composed of a residual connection and a dense block. Due to residual connections, the performance of the network is improved by higher effectiveness of the training stage . On the other hand, dense blocks exploit new features from the received volume. The LUNA16 dataset was used as training data and 10-fold cross-validation was performed for validation. With the highest sensitivity, eight false positives were produced on average per scan.
Ding et al. (86) used a modified Faster R-CNN (after Region-Based Convolutional Neural Network) with stacked deconvolution layers at the end of its standard architecture. CT volume is passed to the network sequentially, with an input image of 600×600×3 voxels from 3 consecutive slices of the series passed to generate region proposals for that subspace of the CT scan. An additional 3D CNN was designed to reduce false positives in the second stage of the pipeline. A LUNA16 dataset was used for performance evaluation. There were an average of eight false positives per scan.
Xie et al. (87) applied a similar approach to identify and classify the lesions. The candidate nodules were selected by a network in three consecutive steps. They used Faster R-CNN with two region proposal networks trained for different kind of slices to detect the nodule. Then additional 2D CNNs were implemented to minimize false positives in the nodule classification. The last network, serving as the voting node, was used for result fusion. Sixty percent of the LUNA16 dataset was used as training data, whereas the remaining data were used as the validation and testing sets. The highest sensitivity was obtained with eight false positives per scan. Huang et al. (88) took advantage of performing data augmentation on training data. They used standard 3D CNN architecture; however, by using smart data augmentation, the classification performance was significantly improved. Each nodule was disturbed randomly, thus generating several variants that enhanced the training set. Experiments were carried out on an LIDC dataset with 10-fold cross-validation. Only five false positives per scan were produced with a sensitivity of 90%. Nasrullah et al. (89) used a Faster R-CNN with customized mixed link network (CMixNet) and U-net-like architecture for nodule detection. The volumetric CT image was divided into 96×96×96 voxel sub-volumes and processed separately; the resulting nodule detection system combined all processed patches. A sensitivity of 94.21% was achieved with an average of eight false positives per scan for the LIDC dataset. Following the idea of R-CNN, Cai et al. (90), used Mask R-CNN with ResNet50 architecture as a backbone and applied a feature pyramid network (FPN) to extract feature maps. Then, a region proposal network (RPN) was used to generate bounding boxes for candidate nodules from the generated feature maps. For the LUNA16 dataset, the proposed method achieved a sensitivity of 88.70% with eight false positives per scan.
Table 4 presents a summary of deep learning algorithms used in the detection and segmentation of pulmonary nodules.

Full table
Deep learning-based automated classification of pulmonary nodules
Automatically detected pulmonary nodules need to be diagnosed in order to determine whether they are malignant or benign. This process, which was originally performed by CAD systems is now a task for deep neural networks. Next, we will discuss the state-of-the-art solutions for such diagnostic algorithms.
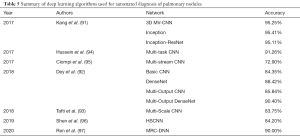
Kang et al., (91) used the 3D multi-view CNN (MV-CNN) based on 3D Inception and 3D Inception-ResNet architectures. ResNet architecture addresses the vanishing gradient problem in deep CNN networks. During backpropagation, the gradient value decreases after each convolution block and barely influences the initial layers in a network. ResNet makes use of the residual connection that short convolution blocks during backpropagation. Inception architecture enhances the convolution operation by introducing a different convolution resolution. The proposed network operates on cropped nodules from the 3D volume at different scales, thus capturing the localized as well as the region-based phenotype of the nodule. Each series of the cropped nodule is fed into the network simultaneously to generate a diagnosis. Experiments were conducted on an LIDC-IDRI dataset with 10-fold cross-validation. Dey et al. (92) investigated the performance of different 3D architectures for nodule classification. Four architectures have been proposed: basic 3D CNN, 3D DenseNet, multi-output CNN, and multi-output DenseNet. Each architecture is composed of two paths, which accept input volumes 50×50×5 voxels and 100×100×10 voxels, respectively. The input image is created by resizing the segmentation to a particular input shape, and then the slices are sampled from the segmented regions. Basic 3D CNN represents a vanilla CNN architecture extended to a 3D task, whereas 3D DenseNet is equipped with additional dense layers before each convolution layer. Multi-output architectures take advantage of early outputs after each pooling layer, which are passed to the final output to improve classification performance. The LIDC-IDRI dataset together with a private dataset of 147 CT scans were used in the study. For the LIDC-IDRI dataset, five-fold cross-validation was used for evaluation.
Taffti et al. (93) used full CT scans to classify nodules into cancerous and non-cancerous groups. In their research, a 3D CNN was constructed by creating three convolution paths for different volume resolutions. Each CT image was resized to three different resolutions (50×50×20, 100×100×20, and 150×150×20 voxels) and fed into the network. Taffti et al.’s study used Data Science Bowl 2017 (DSB2017) and Marshfield Clinic Lung Image Archive datasets. Hussein et al. (94) also used a standard 3D CNN architecture, although the model was pre-trained on 1,000,000 videos. In their work, they introduced the use of transfer learning for lung nodule classification for the first time. Furthermore, the following six separate CNN networks were created to assess different nodule features: calcification, lobulation, sphericity, speculation, margin, and texture. The features were defined by a sparse matrix representation of the first convolution layer in each network. Using these features, a malignancy score for a nodule was produced. An LIDC dataset was used and the performance was evaluated using 10-fold cross-validation.
Ciompi et al. (95) presented a deep learning system for classification based on multi-stream, multi-scale CNN. The system was trained on the data from the Multicentric Italian Lung Detection (MILD) screening trial and tested on images from a Danish Lung Cancer Screening Trial (DLSCT). In the study, the radiomic features of the nodule were linked with classification performance of the system. The lowest classification scores were achieved for solid and part-solid nodules (63.3% and 64.7% respectively), and the highest classification score (89.2%) was obtained for calcified nodules.
Shen (96) developed a deep learning model that produces additional information to form an interpretable framework for assessing nodule malignancy. Apart from diagnosing malignancy, the proposed hierarchical semantic CNN (HSCNN) predicts five different categories: calcification, margin, texture, sphericity, and subtlety. The motivation for this approach was to approach a “black-box” criticism of deep neural networks by producing a set of categories which attempt to explain a classification outcome. An LIDC dataset was used for training and validation. Recently, Ren et al. (97) developed a manifold regularized classification deep neural network (MRC-DNN) with an encoder-decoder structure that outputs a reconstructed image of the volumetric image of an input nodule. During the process, a manifold representation of the nodule is created. By feeding this representation into a fully connected neural network (FCNN), a classification is performed directly on the manifold. The network was trained and validated on an LIDC dataset.
Table 5 presents the summary for the DL systems for automated diagnosis of pulmonary nodules.
Full table
Conclusions
The issue of automated detection (segmentation) of pulmonary nodules and their later diagnosis is still not completely resolved. A number of computer-aided detection systems, as well as radiomic and deep learning approaches exist; however, the gold standard for such methods has yet to be established. Among all the methods discussed here, those based on radiomics and deep learning seem to be the most promising. We are certain that in the not too distant future we will see a successful combination of radiomics and deep learning that will result in a robust, sensitive, and accurate computer-aided diagnostic tool for radiologists.
Acknowledgments
Funding: This work was partially financially supported by the National Science Centre, Poland, OPUS grant no. 2017/27/B/NZ7/01833 (FB, WP, JP).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Witold Rzyman) for the series “Implementation of CT-based screening of lung cancer” published in Translational Lung Cancer Research. The article was sent for external peer review organized by the Guest Editor and the editorial office.
Peer Review File: Available at http://dx.doi.org/10.21037/tlcr-20-708
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-708). The series “Implementation of CT-based screening of lung cancer” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- World Health Organization. Available online: . Accessed 10 May 2020http://www.who.int/en/
- Foy M, Yip R, Chen X, et al. Modeling the mortality reduction due to computed tomography screening for lung cancer. Cancer 2011;117:2703-8. [Crossref] [PubMed]
- Walter JE, Heuvelmans MA, de Jong PA, et al. Occurrence and lung cancer probability of new solid nodules at incidence screening with low-dose CT: analysis of data from the randomised, controlled NELSON trial. Lancet Oncol 2016;17:907-16. [Crossref] [PubMed]
- Oudkerk M, Devaraj A, Vliegenthart R, et al. European position statement on lung cancer screening. Lancet Oncol 2017;18:e754-66. [Crossref] [PubMed]
- Wood DE, Kazerooni EA, Baum SL, et al. Lung cancer screening, version 3.2018, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 2018;16:412-41. [Crossref] [PubMed]
- Attili AK, Kazerooni EA. Imaging of the Solitary Pulmonary Nodule. In Evidence-Based Imaging 2006 (pp. 417-440). Springer, New York, NY.
- Sharma SK, Mohan A. Solitary pulmonary nodule: How and how much to investigate? Med Update 2008;18:824-32.
- Brandman S, Ko JP. Pulmonary nodule detection, characterization, and management with multidetector computed tomography. J Thorac Imaging 2011;26:90-105. [Crossref] [PubMed]
- de Hoop B, Gietema H, van de Vorst S, et al. Pulmonary ground-glass nodules: increase in mass as an early indicator of growth. Radiology 2010;255:199-206. [Crossref] [PubMed]
- Hamet P, Tremblay J. Artificial intelligence in medicine. Metabolism 2017;69S:S36-40. [Crossref] [PubMed]
- Shi Z, Ma J, Zhao M, et al. Many is better than one: an integration of multiple simple strategies for accurate lung segmentation in CT Images. BioMed Res Int 2016;2016:1480423. [Crossref] [PubMed]
- Law TY, Heng P. Automated extraction of bronchus from 3D CT images of lung based on genetic algorithm and 3D region growing. In Medical Imaging 2000: Image Processing 2000 (Vol. 3979, pp. 906-916). International Society for Optics and Photonics.
- Saita S, Kubo M, Kawata Y, et al. An algorithm for the extraction of pulmonary fissures from low‐dose multislice CT image. Systems and Computers in Japan. 2006;37:63-76. [Crossref]
- Lai J, Ye M. Active contour based lung field segmentation. In 2009 International Conference on Intelligent Human-Machine Systems and Cybernetics 2009 (Vol. 1, pp. 288-291). IEEE.
- Zhang Y, Osanlouy M, Clark AR, et al. Pulmonary lobar segmentation from computed tomography scans based on a statistical finite element analysis of lobe shape. In Medical Imaging 2019: Image Processing 2019 (Vol. 10949, p. 1094932). International Society for Optics and Photonics.
- Fischer AM, Varga-Szemes A, Martin SS, et al. Artificial Intelligence-based Fully Automated Per Lobe Segmentation and Emphysema-quantification Based on Chest Computed Tomography Compared With Global Initiative for Chronic Obstructive Lung Disease Severity of Smokers. J Thorac Imaging 2020;35 Suppl 1:S28-S34. [Crossref] [PubMed]
- Campadelli P, Casiraghi E, Artioli D. A fully automated method for lung nodule detection from postero-anterior chest radiographs. IEEE Trans Med Imaging 2006;25:1588-603. [Crossref] [PubMed]
- Gao Q, Wang S, Zhao D, et al. Accurate lung segmentation for X-ray CT images. In Third International Conference on Natural Computation (ICNC 2007) 2007 (Vol. 2, pp. 275-279). IEEE.
- Dai S, Lu K, Dong J, et al. A novel approach of lung segmentation on chest CT images using graph cuts. Neurocomputing 2015;168:799-807. [Crossref]
- Soliman A, Khalifa F, Elnakib A, et al. Accurate lungs segmentation on CT chest images by adaptive appearance-guided shape modeling. IEEE Trans Med Imaging 2017;36:263-76. [Crossref] [PubMed]
- Rebouças Filho PP, Cortez PC, da Silva Barros AC, et al. Novel and powerful 3D adaptive crisp active contour method applied in the segmentation of CT lung images. Med Image Anal 2017;35:503-16. [Crossref] [PubMed]
- Akram S, Javed MY, Akram MU, et al. Pulmonary nodules detection and classification using hybrid features from computerized tomographic images. J Med Imaging Health Inform 2016;6:252-9. [Crossref]
- Pu J, Paik DS, Meng X, et al. Shape “break-and-repair” strategy and its application to automated medical image segmentation. IEEE Trans Vis Comput Graph 2011;17:115-24. [Crossref] [PubMed]
- Jo HH, Hong H, Goo JM. Pulmonary nodule registration in serial CT scans using global rib matching and nodule template matching. Comput Biol Med 2014;45:87-97. [Crossref] [PubMed]
- Kubota T, Jerebko AK, Dewan M, et al. Segmentation of pulmonary nodules of various densities with morphological approaches and convexity models. Med Image Anal 2011;15:133-54. [Crossref] [PubMed]
- Ye X, Lin X, Dehmeshki J, et al. Shape-based computer-aided detection of lung nodules in thoracic CT images. IEEE Trans Biomed Eng 2009;56:1810-20. [Crossref] [PubMed]
- Naqi SM, Sharif M, Yasmin M, et al. Lung nodule detection using polygon approximation and hybrid features from CT images. Curr Med Imaging 2018;14:108-17. [Crossref]
- Huidrom R, Chanu YJ, Singh KM. Pulmonary nodule detection on computed tomography using neuro-evolutionary scheme. Signal Image Video Process 2019;13:53-60. [Crossref]
- Ozekes S, Osman O, Ucan ON. Nodule detection in a lung region that's segmented with using genetic cellular neural networks and 3D template matching with fuzzy rule based thresholding. Korean J Radiol 2008;9:1-9. [Crossref] [PubMed]
- Cascio D, Magro R, Fauci F, et al. Automatic detection of lung nodules in CT datasets based on stable 3D mass–spring models. Comput Biol Med 2012;42:1098-109. [Crossref] [PubMed]
- Soltaninejad S, Keshani M, Tajeripour F. Lung nodule detection by KNN classifier and active contour modelling and 3D visualization. In The 16th CSI International Symposium on Artificial Intelligence and Signal Processing (AISP 2012) 2012 (pp. 440-445). IEEE.
- Choi WJ, Choi TS. Automated pulmonary nodule detection system in computed tomography images: A hierarchical block classification approach. Entropy 2013;15:507-23. [Crossref]
- Gonçalves L, Novo J, Campilho A. Hessian based approaches for 3D lung nodule segmentation. Expert Syst Appl 2016;61:1-15. [Crossref]
- Guo W, Wei Y, Zhou H, et al. An adaptive lung nodule detection algorithm. In 2009 Chinese Control and Decision Conference 2009 (pp. 2361-2365). IEEE.
- Murphy K, van Ginneken B, Schilham AM, et al. A large-scale evaluation of automatic pulmonary nodule detection in chest CT using local image features and k-nearest-neighbour classification. Med Image Anal 2009;13:757-70. [Crossref] [PubMed]
- Retico A, Fantacci ME, Gori I, et al. Pleural nodule identification in low-dose and thin-slice lung computed tomography. Comput Biol Med 2009;39:1137-44. [Crossref] [PubMed]
- Sousa JR, Silva AC, de Paiva AC, et al. Methodology for automatic detection of lung nodules in computerized tomography images. Comput Methods Programs Biomed 2010;98:1-14. [Crossref] [PubMed]
- Messay T, Hardie RC, Rogers SK. A new computationally efficient CAD system for pulmonary nodule detection in CT imagery. Med Image Anal 2010;14:390-406. [Crossref] [PubMed]
- Orozco HM, Villegas OO, Domínguez HD, et al. Lung nodule classification in CT thorax images using support vector machines. In 12th Mexican International Conference on Artificial Intelligence 2013 Nov 24 (pp. 277-283). IEEE.
- Tartar A, Kilic N, Akan A. Classification of pulmonary nodules by using hybrid features. Comput Math Methods Med 2013;2013:148363. [Crossref] [PubMed]
- Teramoto A, Fujita H, Takahashi K, et al. Hybrid method for the detection of pulmonary nodules using positron emission tomography/computed tomography: a preliminary study. Int J Comput Assist Radiol Surg 2014;9:59-69. [Crossref] [PubMed]
- Gong J, Liu JY, Wang LJ, et al. Automatic detection of pulmonary nodules in CT images by incorporating 3D tensor filtering with local image feature analysis. Phys Med 2018;46:124-33. [Crossref] [PubMed]
- Sun L, Wang Z, Pu H, et al. Spectral analysis for pulmonary nodule detection using the optimal fractional S-Transform. Comput Biol Med 2020;119:103675. [Crossref] [PubMed]
- Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. [Crossref] [PubMed]
- Kumar V, Gu Y, Basu S, et al. Radiomics: the process and the challenges. Magn Reson Imaging 2012;30:1234-48. [Crossref] [PubMed]
- Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. Corrected in Nat Commun. 2014;5:4644. [Crossref] [PubMed]
- Cuocolo R, Stanzione A, Ponsiglione A, et al. Clinically significant prostate cancer detection on MRI: A radiomic shape features study. Eur J Radiol 2019;116:144-9. [Crossref] [PubMed]
- Limkin EJ, Reuzé S, Carré A, et al. The complexity of tumor shape, spiculatedness, correlates with tumor radiomic shape features. Sci Rep 2019;9:4329. [Crossref] [PubMed]
- Shafiq-Ul-Hassan M, Zhang GG, Latifi K, et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med Phys 2017;44:1050-62. [Crossref] [PubMed]
- Mackin D, Fave X, Zhang L, et al. Harmonizing the pixel size in retrospective computed tomography radiomics studies. PLoS One 2017;12:e0178524. [Crossref] [PubMed]
- Chung AG, Khalvati F, Shafiee MJ, et al. Prostate cancer detection via a quantitative radiomics-driven conditional random field framework. IEEE Access 2015;3:2531-41.
- Yu H, Scalera J, Khalid M, et al. Texture analysis as a radiomic marker for differentiating renal tumors. Abdom Radiol (NY) 2017;42:2470-8. [Crossref] [PubMed]
- M. D. Anderson Cancer Center Head and Neck Quantitative Imaging Working Group. Investigation of radiomic signatures for local recurrence using primary tumor texture analysis in oropharyngeal head and neck cancer patients. Sci Rep 2018;8:1524. [Crossref] [PubMed]
- Yasaka K, Akai H, Mackin D, et al. Precision of quantitative computed tomography texture analysis using image filtering. Medicine (Baltimore) 2017;96:e6993. [Crossref] [PubMed]
- Cusumano D, Dinapoli N, Boldrini L, et al. Fractal-based radiomic approach to predict complete pathological response after chemo-radiotherapy in rectal cancer. Radiol Med 2018;123:286-95. [Crossref] [PubMed]
- Galloway MM. Texture analysis using grey level run lengths. Computer Graphics and Image Processing 1974;4:172-9. [Crossref]
- Pentland AP. Fractal-based description of natural scenes. IEEE Trans Pattern Anal Mach Intell 1984;6:661-74. [Crossref] [PubMed]
- Amadasun M, King R. Textural features corresponding to textural properties. IEEE Trans Syst Man Cybern 1989;19:1264-74. [Crossref]
- Thibault G, Angulo J, Meyer F. Advanced statistical matrices for texture characterization: application to cell classification. IEEE Trans Biomed Eng 2014;61:630-7. [Crossref] [PubMed]
- Leijenaar RT, Nalbantov G, Carvalho S, et al. The effect of SUV discretization in quantitative FDG-PET Radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep 2015;5:11075. [Crossref] [PubMed]
- van Velden FH, Kramer GM, Frings V, et al. Repeatability of radiomic features in non-small-cell lung cancer (18F) FDG-PET/CT studies: impact of reconstruction and delineation. Mol Imaging Biol 2016;18:788-95. [Crossref] [PubMed]
- Zhao B, Tan Y, Tsai WY, et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep 2016;6:23428. [Crossref] [PubMed]
- Zhovannik I, Bussink J, Traverso A, et al. Learning from scanners: Bias reduction and feature correction in radiomics. Clin Transl Radiat Oncol 2019;19:33-8. [Crossref] [PubMed]
- Yip SS, Aerts HJ. Applications and limitations of radiomics. Phys Med Biol 2016;61:R150-66. [Crossref] [PubMed]
- Lambin P, Leijenaar RT, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017;14:749. [Crossref] [PubMed]
- Parmar C, Grossmann P, Bussink J, et al. Machine Learning methods for Quantitative Radiomic Biomarkers. Sci Rep 2015;5:13087. [Crossref] [PubMed]
- Ferreira JR Junior, Koenigkam-Santos M, Cipriano FEG, et al. Radiomics-based features for pattern recognition of lung cancer histopathology and metastases. Comput Methods Programs Biomed 2018;159:23-30. [Crossref] [PubMed]
- Tajbakhsh N, Suzuki K. Comparing two classes of end-to-end machine-learning models in lung nodule detection and classification: MTANNs vs. CNNs. Pattern Recognit 2017;63:476-86. [Crossref]
- Hosny A, Parmar C, Coroller TP, et al. Deep learning for lung cancer prognostication: A retrospective multi-cohort radiomics study. PLoS Med 2018;15:e1002711. [Crossref] [PubMed]
- National Lung Screening Trial Research Team. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409. [Crossref] [PubMed]
- Kumar D, Chung AG, Shaifee MJ, et al. Discovery radiomics for pathologically-proven computed tomography lung cancer prediction. In International Conference Image Analysis and Recognition 2017 (pp. 54-62). Springer, Cham.
- Liu Y, Balagurunathan Y, Atwater T, et al. Radiological image traits predictive of cancer status in pulmonary nodules. Clin Cancer Res 2017;23:1442-9. [Crossref] [PubMed]
- Wu W, Parmar C, Grossmann P, et al. Exploratory study to identify radiomics classifiers for lung cancer histology. Front Oncol 2016;6:71. [Crossref] [PubMed]
- Chaddad A, Desrosiers C, Toews M, et al. Predicting survival time of lung cancer patients using radiomic analysis. Oncotarget 2017;8:104393. [Crossref] [PubMed]
- Wu J, Aguilera T, Shultz D, et al. Early-stage non–small cell lung cancer: quantitative imaging characteristics of 18F fluorodeoxyglucose PET/CT allow prediction of distant metastasis. Radiology 2016;281:270-8. [Crossref] [PubMed]
- Coroller TP, Grossmann P, Hou Y, et al. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol 2015;114:345-50. [Crossref] [PubMed]
- Aerts HJ, Grossmann P, Tan Y, et al. Defining a radiomic response phenotype: a pilot study using targeted therapy in NSCLC. Sci Rep 2016;6:33860. [Crossref] [PubMed]
- Coroller TP, Agrawal V, Narayan V, et al. Radiomic phenotype features predict pathological response in non-small cell lung cancer. Radiother Oncol 2016;119:480-6. [Crossref] [PubMed]
- Mattonen SA, Tetar S, Palma DA, et al. Automated texture analysis for prediction of recurrence after stereotactic ablative radiation therapy for lung cancer. Int J Radiat Oncol Biol Physics 2015;93:S5-6. [Crossref]
- Bogowicz M, Vuong D, Huellner MW, et al. CT radiomics and PET radiomics: ready for clinical implementation? Q J Nucl Med Mol Imaging 2019;63:355-70. [Crossref] [PubMed]
- Vaidya P, Bera K, Gupta A, et al. CT derived radiomic score for predicting the added benefit of adjuvant chemotherapy following surgery in stage I, II resectable non-small cell lung cancer: a retrospective multicohort study for outcome prediction. Lancet Digital Health 2020;2:e116-28. [Crossref] [PubMed]
- Avanzo M, Stancanello J, Pirrone G, et al. Radiomics and deep learning in lung cancer. Strahlenther Onkol. 2020;196:879-87. [Crossref] [PubMed]
- Khosravan N, Bagci U. S4ND: single-shot single-scale lung nodule detection. In International Conference on Medical Image Computing and Computer-Assisted Intervention 2018 (pp. 794-802). Springer, Cham.
- Golan R, Jacob C, Denzinger J. Lung nodule detection in CT images using deep convolutional neural networks. In 2016 International Joint Conference on Neural Networks (IJCNN) 2016 (pp. 243-250). IEEE.
- Zhu W, Liu C, Fan W, et al. Deeplung: Deep 3d dual path nets for automated pulmonary nodule detection and classification. In 2018 IEEE Winter Conference on Applications of Computer Vision (WACV) 2018 (pp. 673-681). IEEE.
- Ding J, Li A, Hu Z, et al. Accurate pulmonary nodule detection in computed tomography images using deep convolutional neural networks. In International Conference on Medical Image Computing and Computer-Assisted Intervention 2017 (pp. 559-567). Springer, Cham.
- Xie H, Yang D, Sun N, et al. Automated pulmonary nodule detection in CT images using deep convolutional neural networks. Pattern Recognition 2019;85:109-19. [Crossref]
- Huang X, Shan J, Vaidya V. Lung nodule detection in CT using 3D convolutional neural networks. In 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017) 2017 (pp. 379-383). IEEE.
- Nasrullah N, Sang J, Alam MS, et al. Automated lung nodule detection and classification using deep learning combined with multiple strategies. Sensors. 2019;19:3722. [Crossref] [PubMed]
- Cai L, Long T, Dai Y, et al. Mask R-CNN-Based Detection and Segmentation for Pulmonary Nodule 3D Visualization Diagnosis. IEEE Access. 2020;8:44400-9. [Crossref]
- Kang G, Liu K, Hou B, et al. 3D multi-view convolutional neural networks for lung nodule classification. PLoS One 2017;12:e0188290. [Crossref] [PubMed]
- Dey R, Lu Z, Hong Y. Diagnostic classification of lung nodules using 3D neural networks. In 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018) 2018 (pp. 774-778). IEEE.
- Tafti AP, Bashiri FS, LaRose E, et al. Diagnostic classification of lung CT images using deep 3d multi-scale convolutional neural network. In 2018 IEEE International Conference on Healthcare Informatics (ICHI) 2018 (pp. 412-414). IEEE.
- Hussein S, Cao K, Song Q, et al. Risk stratification of lung nodules using 3D CNN-based multi-task learning. In International conference on information processing in medical imaging 2017 (pp. 249-260). Springer, Cham.
- Ciompi F, Chung K, Van Riel SJ, et al. Towards automatic pulmonary nodule management in lung cancer screening with deep learning. Sci Rep 2017;7:46479. [Crossref] [PubMed]
- Shen S, Han SX, Aberle DR, et al. An interpretable deep hierarchical semantic convolutional neural network for lung nodule malignancy classification. Expert Syst Appl 2019;128:84-95. [Crossref] [PubMed]
- Ren Y, Tsai MY, Chen L, et al. A manifold learning regularization approach to enhance 3D CT image-based lung nodule classification. Int J Comput Assist Radiol Surg 2020;15:287-95. [Crossref] [PubMed]




