
Concomitant mutation status of ALK -rearranged non-small cell lung cancers and its prognostic impact on patients treated with crizotinib
Introduction
Lung cancer remains the most common malignancy worldwide, the morbidity and mortality of which are at the forefront of global research (1). With the recent development in targeted therapy, treatment for patients with non-small cell lung cancer (NSCLC) has changed substantially (2). Molecular studies have shown that 64% of lung adenocarcinomas have driver gene alterations (3).
Similar to that of Kirsten rat sarcoma viral oncogene homolog (KRAS) and epidermal growth factor receptor (EGFR), the discovery of echinoderm microtubule-associated protein-like 4 fused with anaplastic lymphoma kinase (EML4-ALK) has been hugely significant to the individualized treatment of NSCLC. The ALK gene encodes a transmembrane tyrosine kinase receptor. EML4-ALK translocation can result in constitutive ALK kinase activity and represents an oncogenic addiction pathway in lung cancer. EML4-ALK fusion protein serves as a therapeutic target for an ALK-TKI, and has shown promising results when used to treat NSCLC patients carrying ALK rearrangement. Over the last few years, ALK inhibitors, including the TKI crizotinib, have shown significant benefits in the management of ALK-positive NSCLC compared to conventional chemotherapy (4). Of patients with NSCLC, approximately 3–7% express EML4-ALK and can benefit from individualized treatment (5). There is a subset of ALK rearrangement-positive NSCLCs that respond differently to ALK inhibitors according to variations in EML4-ALK fusion. EML4-ALK fusion variant V3 is a high-risk feature in ALK-positive NSCLC and confers early metastatic spread, treatment failure after tyrosine kinase inhibitor (TKI) therapy, chemotherapy, and cerebral radiotherapy, as well as inferior overall survival (OS) (6). However, it has been reported that there is no correlation between variants of EML4-ALK and patients’ clinical responses to crizotinib (3). Although several ALK inhibitors, such as crizotinib, ceritinib, alectinib, and brigatinib, have been approved for cancer therapy, a large number of NSCLC patients go on to develop disease progression after the application of ALK TKIs (7). The clinicopathological features of patients with both ALK fusion and an oncogene mutation are critical to progressing individualized treatment for NSCLC (8).
So far, crizotinib has the longest follow-up of any drug for the treatment of ALK+ NSCLC, as it was the first drug to be approved (9). However, most patients who respond to frontline TKIs eventually acquire resistance within 1–2 years (10). According to published research reports, the mechanisms of ALK-TKI resistance can be roughly divided into 2 main forms: ALK-dependent changes and ALK-independent changes. ALK-dependent changes include secondary ALK mutations and ALK gene amplification, whereas ALK-independent changes mainly consist of the upregulation of a bypass signaling pathway and changes in lineages. Among the genetic factors, co-alterations of ALK fusions and gene mutations are the leading cause of crizotinib resistance. Preclinical data have shown that various concomitant mutations (11,12) are associated with low sensitivity to crizotinib (13). In terms of the molecular pathology, rearrangement of ALK is independent of EGFR and KRAS mutations (14-18), although these alterations are not absolutely mutually exclusive (18,19). For instance, in a recent lung cancer mutant complex series, 8% of ALK-positive adenocarcinoma patients also had EGFR or KRAS mutations (20). Studies have illustrated that patients with concomitant EGFR mutation and EML4-ALK translocation are insensitive to EGFR TKIs but sensitive to crizotinib (21). KRAS mutations are the most common concomitant mutations with EML4-ALK fusions, accounting for approximately 2.5% of patients with EML4-ALK NSCLC, and are associated with reduced reactivity to crizotinib and a poor prognosis (22).
In the present study, we analyzed the basic clinical information of 99 patients with ALK-positive NSCLC who were treated in Tianjin Medical University Cancer Institute and Hospital from 2012 to 2016. Next-generation sequencing (NGS) was conducted on 96 paraffin-embedded tissue samples from these patients. According to the NGS results, we analyzed the landscapes of ALK fusion variants and the co-occurring genomic alterations in ALK fusion-positive NSCLC, and explored their associations with patients’ clinical outcomes.
We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/tlcr-21-160).
Methods
Patient information
All patients (n=99) with ALK fusion-positive lung cancer, as assessed by immunohistochemistry [IHC; VENTANA anti-ALK (D5F3) Rabbit Monoclonal Primary Antibody], who received treatment in Tianjin Cancer Hospital between January 2012 and August 2016 were enrolled in the study. Because the samples from the patients were placed for too long, 3 tissue samples failed the quality control of sequencing data, and only 96 samples were tested by NGS for subsequent analysis. The patients’ medical records were retrieved to collect data including clinicopathological characteristics, treatment history, and survival outcomes. Progression-free survival (PFS) after treatment and overall survival (OS) were assessed. This study was approved by the Institutional Review Board of Tianjin Cancer Hospital (No. bc2019089), and was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from all participants.
DNA isolation and capture-based targeted DNA sequencing
DNA was isolated from tumor tissues using a QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany). The NGS library was prepared with a minimum of 50 ng of DNA as described previously (23). Briefly, DNA fragmentation was performed using a Covaris M220 Focused-ultrasonicator (Covaris, MA, USA), after which end repair, phosphorylation, and adapter ligation were carried out. Selective purification of DNA fragments between 200–400 bp was conducted using magnetic beads (Agencourt AMPure XP Kit, Beckman Coulter, CA, USA), followed by hybridization with RNA probes of a panel consisting of 520 cancer-related genes spanning 1.64 megabases (Mb) of the human genome (OncoScreen Plus, Burning Rock Biotech, Guangzhou, China). The targeted library was subsequently enriched by polymerase chain reaction (PCR) amplification. The quality and size of the library were assessed using a Qubit 2.0 Fluorometer with the dsDNA High Sensitivity Assay Kit (Life Technologies, Carlsbad, CA, USA). Sequencing of indexed samples was performed on the NextSeq 500 platform (Illumina, Inc., USA) with paired-end reads and a median sequencing depth of 1,636×.
Data analysis
Sequencing data in FASTQ format were aligned to the reference human genome (hg19) using Burrows–Wheeler Aligner v.0.7.10 (24). Local alignment optimization, duplication marking, and variant calling were conducted using the Genome Analysis Tool Kit v.3.2 (25) and VarScan v.2.4.3 (26). Variants with a loci depth <100 were filtered out with the VarScan fpfilter pipeline. Variants with population frequencies over 0.1% in the Exome Aggregation Consortium (ExAC), 1,000 Genomes, Single Nucleotide Polymorphism Database (dbSNP), or ESP6500SI-V2 databases were excluded from further analysis. The remaining variants were annotated with ANNOtate VARiation (ANNOVAR) (February 1, 2016, release) (27) and SnpEff v.3.6. (28) DNA translocation and copy number variation (CNV) were analyzed using Factera v.1.4.3 (29) and an in-house algorithm based on sequencing depth, respectively. Fusions, large genomic rearrangements, and mutations occurring on the kinase domains of EGFR and ALK were excluded from the mutation count.
Statistical analysis
Statistical analyses were performed using R version 3.3.3 software. Differences among groups were calculated by Fisher’s exact test, paired two-tailed Student’s t-test, or analysis of variance, as appropriate. Survival outcomes were estimated with Kaplan-Meier curves, and the log-rank test was applied to determine the difference in the survival curves between groups. A P value <0.05 was considered to be statistically significant.
Results
Clinicopathological characteristics of the patients
Qualified sequencing data were generated from tissue samples from 96 of the 99 patients and used in subsequent analyses. Table 1 details the clinicopathological characteristics of the 96 patients. The median age of this cohort was 56.5 years, and 52 (54.2%) patients were male. Twenty-nine (30.2%) patients had a history of smoking. There were 81 (84.4%) and 9 (9.4%) diagnoses of lung adenocarcinoma and squamous carcinoma, respectively. Six patients had pulmonary tumors with other histologies (1 adenosquamous carcinoma, 1 mucoepidermoid carcinoma, 1 typical carcinoid, 1 atypical carcinoid, and 2 large cell neuroendocrine carcinoma). There were 31 (32.3%), 17 (17.7%), 28 (29.2%), and 20 (20.8%) patients in stage I, II, III, and IV, respectively. Brain, bone, and visceral metastases were reported in 14 (14.6%), 9 (9.4%), and 7 (7.3%) patients, respectively. A total of 39 (40.6%) patients received crizotinib, 26 of whom were in stage III or IV. Of the 39 patients who were treated with crizotinib, there was prognostic information available for 37. A median progression-free survival (mPFS) of 12 months and a median overall survival (mOS) of 23 months were observed. Patients treated with crizotinib had a poorer prognosis than untreated patients, which was attributable to crizotinib-treated patients having more advanced tumors.

Full table
The genomic landscape of the cohort
Qualified sequencing data were generated from 96 of the 99 patients. NGS identified ALK fusions in 83% (80/96) of patients, and 1 patient harbored concomitant ALK p.S267I mutation and amplification. Among the ALK-rearranged tumors, we identified 4 adenocarcinomas with other concurrent driver mutations: 2 KRAS G12, 1 EGFR 19del, and 1 MET exon14 skipping (Figure 1). The most frequently mutated gene in this cohort was TP53 (24%), followed by lysine (K)-specific methyltransferase 2C (KMT2C; 12%), SET domain containing 2 (SETD2; 10%), and telomerase reverse transcriptase (TERT; 8%). The other concomitant alterations are illustrated in Figure 1.
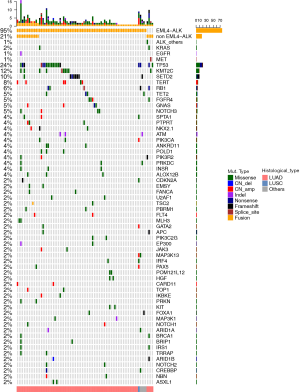
The distribution of ALK-fusion variants
Of the 80 ALK fusion-positive patients, detected with both NGS and IHC, 63 (78.8%) harbored a single EML4-ALK fusion, and 13 (16.2%) patients harbored more than 1 ALK fusion, with EML4-ALK being 1 of the variants. Four (5%) patients carried only non-EML4-ALK fusions (Figure 2A).
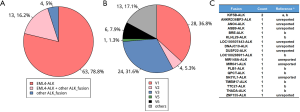
Among the 76 patients carrying EML4-ALK, variant 1 (36.8%) and variant 3 (31.6%) were the most common variants, followed by variants 4 (17.1%), 6 (7.89%), 2 (5.26%), and 5 (1.32%). Thirteen patients harbored EML-ALK variants other than variants 1–6 (Figure 2B). Overall, 19 non-EML4-ALK partners, including 10 novel partners, were identified (Figure 2C).
The prognostic value of ALK–fusion variants and concomitant genomic alterations in patients treated with crizotinib
To identify potential prognostic biomarkers, we investigated the correlations of ALK-fusion variants and concomitant genomic alterations with PFS and OS in patients treated with crizotinib. Patients harboring EML4-ALK showed no significant difference in PFS (P=0.51) (95% CI: 0.429–5.493) or OS (P=0.33) (95% CI: 0.447–11.413) when compared with those carrying non-EML4-ALK partners. Also, the PFS and OS of patients with EML4-ALK variant 3 were comparable to those of patients harboring EML4-ALK variant 1 [P=0.47 (95% CI: 0.389–7.857) and P=0.49 (95% CI: 0.339–9.232)] or other non-V3 fusions [P=0.15 (95% CI: 0.781–4.903) and P=0.37 (95% CI: 0.548–5.121)]. The significance of these results is limited by the small number of patients in our study.
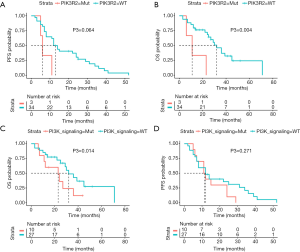
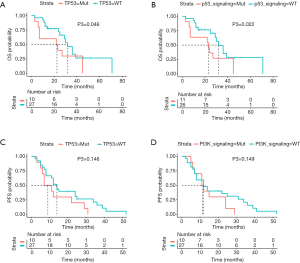
Patients concurrently harboring PIK3R2 alterations showed a trend toward a shorter PFS [6 vs. 13 months, P=0.064 (95% CI: 0.928–14.937), Figure 3A] and significantly shorter OS [11 vs. 32 months, P=0.004 (95% CI: 1.507– 27.643), Figure 3B] than PIK3R2-wild-type patients. We next explored the association of alterations in the PI3K signaling pathway with patient prognosis, and a shorter mOS was observed in patients with these alterations than in patients without them [23 vs. 32 months, P=0.014 (95% CI: 0.072–0.746), Figure 3C]; in contrast, we did not find a significant difference in the PFS of patients with and without alterations (Figure 3D). Furthermore, a shorter mOS was observed in patients harboring concomitant TP53 mutations [22.5 vs. 30 months, P=0.046 (95% CI: 1.023–10.527), Figure 4A] and patients with mutations in the P53 signaling pathway [23 vs. 32 months, P=0.022 (95% CI: 0.729–3.871), Figure 4B] than in patients with TP53/P53-wild-type; however, no difference was found in PFS (Figure 4C,D).
Discussion
In this study, 96 patients with ALK fusions identified by IHC were retrospectively evaluated (Figure S1). Among them, 80 (83%) patients had rearrangement of ALK confirmed by NGS. Also, we discovered 10 novel ALK-fusion partners and identified concurrent mutations that could affect the clinical outcomes to crizotinib therapy. Notably, NGS positivity was correlated with a higher disease control rate and longer PFS than NGS negativity (P=0.02 and P=0.09), while fluorescence in situ hybridization (FISH) and IHC status were not able to distinguish the outcome after treatment with crizotinib. Although it is considered the gold standard, FISH presents a certain false-negative rate. Ventana-D5F3 IHC is qualified as a screening tool, while NGS positivity may more accurately predict clinical benefit of crizotinib, allowing for efficient testing for specific variants and concurrent genomic alterations (30).
The mutual exclusivity of the different driver mutations in adenocarcinoma is generally agreed upon. However, we identified concurrent KRAS G12, EGFR 19del, and MET exon14 skipping in 4 ALK-rearranged adenocarcinomas (4/80; 5.0%). Treatment for this subset of patients remains controversial with respect to the choice of single- vs. dual-targeted therapies (2), and a decision guided by evidence from large-scale studies is still awaited. We also found that 13 patients had other non-classical ALK fusions accompanying the EML4-ALK classic fusion. However, due to the insufficient number of samples, it is impossible to make an effective statistical analysis on the prognosis of these patients, so a large sample of cases is needed to verify it in the future.
Consistent with previously reported studies, we found EML4-ALK fusion to be the most common type of ALK fusion, with variants 1 and variants 3 being the most frequent EML4-ALK variants. Furthermore, we identified 19 different non-EML4-ALK partners, including 10 novel partners (ANKRD36BP2-ALK, ANO4-ALK, ASB9-ALK, LOC100507443-ALK, DNAJC10-ALK, DUSP22-ALK, MIR148A-ALK, MIR641-ALK, SH3YL1-ALK, and ZNF155-ALK) that have not been reported before (31). Although NGS analyses have identified a variety of ALK rearrangements, whether or not the diversity of fusion sites leads to different responses to ALK inhibitors remains unclear. The investigation on the prognostic impacts of ALK-fusion partner/variants in ALK inhibitor-treated patients would guide the accurate stratification of ALK-rearranged NSCLC patients for optimizing the therapeutic selection for individual patients. In line with some other reports (32), but in contrast with other retrospective studies (33,34) and results from the randomized phase 3 trial ALTA-1L (35), we found no significant difference in PFS (P=0.47) or OS (P=0.49) between patients with variant 1 and variant 3 (Figure S2). It is noteworthy that, instead of using DNA-based NGS, most studies that detected a difference in patient outcome according to the EML4-ALK fusion variant used RNA-based NGS (36), which has demonstrated a higher yield for the detection of oncogenic fusions in lung adenocarcinoma (37), or highly sensitive circulating tumor DNA assays, such as ctDx-Lung in case of the ALTA-1L analysis (38). In our study, patients with EML4–ALK variant 3 also had comparable PFS and OS to patients harboring other non-V3 fusions (P=0.15 and P=0.37) (5,33,39,40). A recent study reported that the specific ALK variant seemed to impact the ALK resistance mutations acquired. Further study of the relationship between ALK variants and ALK-TKI-resistant mutations using tissue samples obtained from patients with disease progression is worthy of attention in the future (32).
Besides, in our study, NGS identified TP53 mutations to be the most common type of concomitant mutation with ALK rearrangement, with an incidence of 24%, which was comparable to the 20–29% reported by other investigators (1,41). In addition to identifying the mutations concomitant with ALK rearrangement, we also analyzed the relationships of these combinations with prognosis. Consistent with the findings of other studies, concomitant TP53 mutations were predictive of poor survival (42). Also, we observed that patients with alterations in the PI3K signaling pathway had a shorter mOS than patients without these alterations (23 vs. 32 months, P=0.014). However, due to the small number of cases in the present study, it is necessary to increase the number of samples for further analysis in the future. In line with our findings, another study reported that PI3K/AKT signaling activation led to treatment with ALK inhibitor being ineffective, while PI3K inhibitor increased sensitivity to ALK inhibitor in EML4-ALK-positive cells (43). We hope that further research on the ALK signaling pathway will lead to the emergence of novel generations of ALK inhibitors, which will aid in improving the PFS and OS of patients with ALK rearrangements. Since the incidence of mutations concomitant with ALK rearrangement has been discovered to be high, the relationship between concomitant mutations in the ALK signaling pathway and prognosis needs to be clarified so that the targeted therapy that will maximize the benefit to the patient and substantially improve their survival can be chosen (44).
Limitations of our study which might weaken the strength its findings should also be noted; these include its retrospective nature and the small sample size of patients with ALK rearrangements, especially those receiving ALK inhibitors. Therefore, the prognostic impact of the concomitant alterations we discovered requires confirmation by further prospective studies with large cohorts. Through this, accurate stratification of patients with ALK-rearranged NSCLC for selecting those who may receive the optimal therapeutic benefit may be realized.
Acknowledgments
The authors appreciate the academic support from AME Lung Cancer Collaborative Group.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-21-160
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-21-160
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-21-160). Dr. MGD reports grants from Takeda and BluePrint Medicine, personal fees from Pfizer, BMS, and Boehringer Ingelheim, grants and personal fees from AstraZeneca, outside the submitted work. Dr. PC reports grants and personal fees from Novartis, Roche, AstraZeneca, and Takeda, personal fees from Pfizer, Chugai and Boehringer, outside the submitted work. Dr. AJW reports grants from Astra Zeneca, grants and personal fees from Boehringer Ingelheim, Pfizer, Roche, Takeda, outside the submitted work. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Institutional Review Board of Tianjin Cancer Hospital (No. bc2019089), and was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All patients provided written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Li M, Hou X, Zhou C, et al. Prevalence and Clinical Impact of Concomitant Mutations in Anaplastic Lymphoma Kinase Rearrangement Advanced Non-small-Cell Lung Cancer (Guangdong Association of Thoracic Oncology Study 1055). Front Oncol 2020;10:1216. [Crossref] [PubMed]
- Chen RL, Zhao J, Zhang XC, et al. Crizotinib in advanced non-small-cell lung cancer with concomitant ALK rearrangement and c-Met overexpression. BMC Cancer 2018;18:1171. [Crossref] [PubMed]
- Christopoulos P, Endris V, Bozorgmehr F, et al. EML4-ALK fusion variant V3 is a high-risk feature conferring accelerated metastatic spread, early treatment failure and worse overall survival in ALK(+) non-small cell lung cancer. Int J Cancer 2018;142:2589-98. [Crossref] [PubMed]
- Du X, Shao Y, Qin HF, et al. ALK-rearrangement in non-small-cell lung cancer (NSCLC). Thorac Cancer 2018;9:423-30. [Crossref] [PubMed]
- Lei YY, Yang JJ, Zhang XC, et al. Anaplastic Lymphoma Kinase Variants and the Percentage of ALK-Positive Tumor Cells and the Efficacy of Crizotinib in Advanced NSCLC. Clin Lung Cancer 2016;17:223-31. [Crossref] [PubMed]
- Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693-703. [Crossref] [PubMed]
- Radaram B, Pisaneschi F, Rao Y, et al. Novel derivatives of anaplastic lymphoma kinase inhibitors: Synthesis, radiolabeling, and preliminary biological studies of fluoroethyl analogues of crizotinib, alectinib, and ceritinib. Eur J Med Chem 2019;182:111571 [Crossref] [PubMed]
- Boland JM, Jang JS, Li J, et al. MET and EGFR mutations identified in ALK-rearranged pulmonary adenocarcinoma: molecular analysis of 25 ALK-positive cases. J Thorac Oncol 2013;8:574-81. [Crossref] [PubMed]
- Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014;371:2167-77. [Crossref] [PubMed]
- Katayama R, Lovly CM, Shaw AT. Therapeutic targeting of anaplastic lymphoma kinase in lung cancer: a paradigm for precision cancer medicine. Clin Cancer Res 2015;21:2227-35. [Crossref] [PubMed]
- Katayama R, Shaw AT, Khan TM, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med 2012;4:120ra17 [Crossref] [PubMed]
- Lin JJ, Riely GJ, Shaw AT. Targeting ALK: Precision Medicine Takes on Drug Resistance. Cancer Discov 2017;7:137-55. [Crossref] [PubMed]
- Yun MR, Choi HM, Lee YW, et al. Targeting YAP to overcome acquired resistance to ALK inhibitors in ALK-rearranged lung cancer. EMBO Mol Med 2019;11:e10581 [Crossref] [PubMed]
- Hernández L, Pinyol M, Hernández S, et al. TRK-fused gene (TFG) is a new partner of ALK in anaplastic large cell lymphoma producing two structurally different TFG-ALK translocations. Blood 1999;94:3265-8. [Crossref] [PubMed]
- Sholl LM, Weremowicz S, Gray SW, et al. Combined use of ALK immunohistochemistry and FISH for optimal detection of ALK-rearranged lung adenocarcinomas. J Thorac Oncol 2013;8:322-8. [Crossref] [PubMed]
- Takahashi T, Sonobe M, Kobayashi M, et al. Clinicopathologic features of non-small-cell lung cancer with EML4-ALK fusion gene. Ann Surg Oncol 2010;17:889-97. [Crossref] [PubMed]
- Wong DW, Leung EL, So KK, et al. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer 2009;115:1723-33. [Crossref] [PubMed]
- Zhang X, Zhang S, Yang X, et al. Fusion of EML4 and ALK is associated with development of lung adenocarcinomas lacking EGFR and KRAS mutations and is correlated with ALK expression. Mol Cancer 2010;9:188. [Crossref] [PubMed]
- Camidge DR, Kono SA, Flacco A, et al. Optimizing the detection of lung cancer patients harboring anaplastic lymphoma kinase (ALK) gene rearrangements potentially suitable for ALK inhibitor treatment. Clin Cancer Res 2010;16:5581-90. [Crossref] [PubMed]
- Kris MG, Johnson BE, Kwiatkowski DJJJoCO. Identification of driver mutations in tumor specimens from 1,000 patients with lung adenocarcinoma: The NCI's Lung Cancer Mutation Consortium (LCMC). J Clin Oncol 2011;29:abstr CRA7506.
- Vestergaard HH, Christensen MR, Lassen UN. A systematic review of targeted agents for non-small cell lung cancer. Acta Oncol 2018;57:176-86. [Crossref] [PubMed]
- Ulivi P, Chiadini E, Dazzi C, et al. Nonsquamous, Non-Small-Cell Lung Cancer Patients Who Carry a Double Mutation of EGFR, EML4-ALK or KRAS: Frequency, Clinical-Pathological Characteristics, and Response to Therapy. Clin Lung Cancer 2016;17:384-90. [Crossref] [PubMed]
- Mao X, Zhang Z, Zheng X, et al. Capture-Based Targeted Ultradeep Sequencing in Paired Tissue and Plasma Samples Demonstrates Differential Subclonal ctDNA-Releasing Capability in Advanced Lung Cancer. J Thorac Oncol 2017;12:663-72. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754-60. [Crossref] [PubMed]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. [Crossref] [PubMed]
- Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568-76. [Crossref] [PubMed]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164 [Crossref] [PubMed]
- Cingolani P, Platts A. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80-92. [Crossref] [PubMed]
- Newman AM, Bratman SV, Stehr H, et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics 2014;30:3390-3. [Crossref] [PubMed]
- Lin C, Shi X, Yang S, et al. Comparison of ALK detection by FISH, IHC and NGS to predict benefit from crizotinib in advanced non-small-cell lung cancer. Lung Cancer 2019;131:62-8. [Crossref] [PubMed]
- Camidge DR, Dziadziuszko R, Peters S, et al. Updated Efficacy and Safety Data and Impact of the EML4-ALK Fusion Variant on the Efficacy of Alectinib in Untreated ALK-Positive Advanced Non-Small Cell Lung Cancer in the Global Phase III ALEX Study. J Thorac Oncol 2019;14:1233-43. [Crossref] [PubMed]
- Lin JJ, Zhu VW, Yoda S, et al. Impact of EML4-ALK Variant on Resistance Mechanisms and Clinical Outcomes in ALK-Positive Lung Cancer. J Clin Oncol 2018;36:1199-206. [Crossref] [PubMed]
- Woo CG, Seo S, Kim SW, et al. Differential protein stability and clinical responses of EML4-ALK fusion variants to various ALK inhibitors in advanced ALK-rearranged non-small cell lung cancer. Ann Oncol 2017;28:791-7. [Crossref] [PubMed]
- Christopoulos P, Kirchner M, Endris V, et al. EML4-ALK V3, treatment resistance, and survival: refining the diagnosis of ALK(+) NSCLC. J Thorac Dis 2018;10:S1989-91. [Crossref] [PubMed]
- Camidge DR, Niu H, Kim HR, et al. Correlation of baseline molecular and clinical variables with ALK inhibitor efficacy in ALTA-1L. 2020;38:9517-.
- Volckmar AL, Leichsenring J, Kirchner M, et al. Combined targeted DNA and RNA sequencing of advanced NSCLC in routine molecular diagnostics: Analysis of the first 3,000 Heidelberg cases. Int J Cancer 2019;145:649-61. [Crossref] [PubMed]
- Benayed R, Offin M, Mullaney K, et al. High Yield of RNA Sequencing for Targetable Kinase Fusions in Lung Adenocarcinomas with No Mitogenic Driver Alteration Detected by DNA Sequencing and Low Tumor Mutation Burden. Clin Cancer Res 2019;25:4712-22. [Crossref] [PubMed]
- Supplee JG, Milan MSD, Lim LP, et al. Sensitivity of next-generation sequencing assays detecting oncogenic fusions in plasma cell-free DNA. Lung Cancer 2019;134:96-9. [Crossref] [PubMed]
- Yoshida T, Oya Y, Tanaka K, et al. Differential Crizotinib Response Duration Among ALK Fusion Variants in ALK-Positive Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3383-9. [Crossref] [PubMed]
- Cha YJ, Kim HR, Shim HS. Clinical outcomes in ALK-rearranged lung adenocarcinomas according to ALK fusion variants. J Transl Med 2016;14:296. [Crossref] [PubMed]
- Christopoulos P, Kirchner M, Bozorgmehr F, et al. Identification of a highly lethal V3(+) TP53(+) subset in ALK(+) lung adenocarcinoma. Int J Cancer 2019;144:190-9. [Crossref] [PubMed]
- Wang WX, Xu CW, Chen YP, et al. TP53 mutations predict for poor survival in ALK rearrangement lung adenocarcinoma patients treated with crizotinib. J Thorac Dis 2018;10:2991-8. [Crossref] [PubMed]
- Yang L, Li G, Zhao L, et al. Blocking the PI3K pathway enhances the efficacy of ALK-targeted therapy in EML4-ALK-positive nonsmall-cell lung cancer. Tumour Biol 2014;35:9759-67. [Crossref] [PubMed]
- Christopoulos P, Budczies J, Kirchner M, et al. Defining molecular risk in ALK(+) NSCLC. Oncotarget 2019;10:3093-103. [Crossref] [PubMed]


