
Conventional and semi-automatic histopathological analysis of tumor cell content for multigene sequencing of lung adenocarcinoma
Introduction
Focused multigene testing for druggable genetic aberrations is paramount for personalized therapy of various tumor entities (1), including non-squamous non-small cell lung cancer (NSCLC), particularly, adenocarcinomas (ADC) (2).
Several sequencing technologies have been successfully adapted to analyze formalin-fixed and paraffin-embedded (FFPE) tissue samples and to allow highly sensitive and specific detection of genetic alterations in routine diagnostics. A crucial pre-analytic step is accurate determination of the tumor cell content (TCC) since TCC correlates with variant allele frequencies (VAF) of detected somatic mutations and is therefore important for the interpretation of results. For example, overestimation of TCC can impair valid testing, as VAF of specific mutations may fall below the limit of detection causing false-negative results. Furthermore, an accurate TCC aids inference about the presence of different tumor subclones, supports classification of mutations as somatic vs. germline, and is important for the assessment of sequencing quality metrics (3). Accuracy of TCC quantification becomes even more important when large sequencing panels are employed (4-6). Here, the focus does not only lie on a specific set of targetable driver mutations (hotspots) but also on a comprehensive analysis of a plethora of mutations including subclonal co-mutations with lower VAF. Therefore, reliable mutation calling pipelines are crucial for these approaches but they depend on valid and reliable upfront TCC quantification.
In most diagnostic settings, a pathologist will evaluate the TCC by microscopic inspection of a hematoxylin-eosin (H&E) stained tissue section. However, this approach was shown to suffer from limited interobserver reliability (7-9). Data from previous studies suggest that whereas mean estimates from multiple observers are correct (9) variability among observers is high with a significant proportion overestimating TCC which may lead to false-negative sequencing results (8,10).
Recently, it was demonstrated that digital pathology coupled with image analysis may help to increase accuracy, reproducibility and standardization when evaluating tissue sections (11,12). Commercial as well as open source digital analysis solutions are available, that are able to discern between tumor and stromal cells (13-16).
In the current study we compared conventional (conv) and computer-aided quantification (dPat: digital pathology) of TCC in a large sample set of 120 pulmonary ADC sections. We investigated the influence of several potential confounding variables such as (I) H&E versus immunohistochemical thyroid transcription factor 1 (TTF-1) staining, (II) professional experience of raters, (III) different ADC growth patterns and developed a web-based self-training and quality-improvement tool.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-1168).
Methods
Study design
For a comprehensive evaluation of computer-aided digital TCC quantification in comparison to microscopy-based visual assessment (Figure 1), we compiled a cohort of H&E and TTF-1 stained ADC sections of surgically resected cases. Following digitalization of the sections (20×, Hamamatsu C9600-02 NanoZoomer Digital Pathology, Hamamatsu Photonics K.K., Hamamatsu City, Japan and Aperio CS2, Leica Biosystems, Wetzlar, Germany), 50 images with an area of 1.5 to 2.0 mm2 were chosen independently for H&E staining and immunohistochemistry (IHC), representing 10 cases of each of the five diagnostically relevant histological growth patterns (lepidic, acinar, papillary, solid, micropapillary) (17). In addition, 20 of these images (10 of each staining and 2 of each growth pattern) were duplicated and rotated by 180°, in order to evaluate the intraobserver reliability of the examination. In total 120 images were prepared.
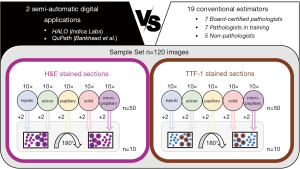
The TCC in each image was determined using two different software applications for digital pathology, the commercially available HALO image analysis platform (18) (Indica Labs, Albuquerque, NM, USA) and the open source software QuPath (13). For the purpose of comparison, the TCC was also estimated by a group of board-certified pathologists (BP, n=7), pathologists in training (PT, n=7) and non-pathologists (NP, n=5) from eight different German pathology institutes.
The images were made available to study participants using a proprietary online tool (www.hd-molpath.de/tcc-test; www.hd-molpath.de/tcc-trainer). The order of the images was once randomized and then kept for all estimators, first showing all the H&E and then the TTF-1 IHC stained images.
Sample material
All samples included in this study were derived from ADC resected at the Thoraxklinik at Heidelberg University. The tumors were diagnosed at the Institute of Pathology at Heidelberg University according to the criteria of the current WHO Classification [2015] for lung cancer (19). The preparation of FFPE samples as well as immuno-/histochemical stainings were supported by the tissue bank of the National Center for Tumor Diseases (NCT; project: #1746, #2015).
H&E staining
H&E stainings were prepared using the Leica Autostainer XL (Leica Biosystems, Nussloch, Germany), Shandon GillTM 3 hematoxylin (Thermo Fisher Scientific, Waltham, MA, USA) and an eosin solution consisting of 7.5 g Eosin G (Merck, Darmstadt, Germany) dissolved in 1,000 mL EtOH (70%) + 5 mL acetic acid (99%). The following successional 1 min incubation steps were performed for the staining reaction: 2× xylene, ethanol 100%, 2× ethanol 96%, ethanol 70%, dH2O, Shandon GillTM 3 hematoxylin, 2× dH2O, ethanol 70%, eosin, ethanol 70%, 2× ethanol 96%, 2× ethanol 100%, 3× xylene.
IHC
IHC for TTF-1 was carried out applying a 1:100 dilution of a mouse monoclonal antibody against TTF-1, clone SPT24 (Leica Biosystems, Newcastle Upon Tyne, UK) to the sections using a BenchMark ULTRA autostainer (Ventana Medical Systems, Tucson, AZ, USA) according to the manufacturer’s instructions.
Tumor annotation, training of tumor and stroma classifier and recording of results
For the semi-automatic digital estimation of the TCC, the digitalized images were analyzed with the two digital pathology software solutions QuPath (13) (v.0.1.2) and HALO (v.2.0.1038, Indica Labs, Albuquerque, NM, USA). Following cell detection using standard parameters, representative areas of tumor and non-neoplastic cells were annotated by a pathologist (MK) for both programms and a tumor-non-neoplastic classifier was trained for each image separately using the default training settings implemented in QuPath and HALO. After training, the whole image was classified and the results were reviewed by a pathologist. If a significant estimated proportion (≥5%) of cells were apparently misclassified during the quality review (either tumor cells as non-neoplastic cells or vice versa), the classifier training was repeated until a correct result was reached. Finally for each section, the absolute number of tumor and non-neoplastic cells were recorded and the TCC as the percentage of tumor cells to total cell number was calculated.
Statistical analysis and plot generation
All statistical analyses were carried out using the R software (v.3.5.0; R Core Team, 2016). The following functions of the “stats” package (v.3.3.0) were used for the stated statistical test: “fisher.test” to perform Fisher’s exact test; “shapiro.test” to perform the Shapiro-Wilk test of normality; “wilcox.test” to perform the Mann-Whitney U test; “cor.test” to test for correlation using Pearson’s product moment correlation coefficient.
Intra-class correlation coefficients (ICC) according to Shrout & Fleiss (20) were calculated using the “icc” function of the “irr” package (v.0.84.1). For comparison of the agreement of the estimators within a group ICC type 2,1 (two-way random, single measures) and for the agreement between the average estimations of different estimator groups ICC type 2,k (two-way random, average measures). Further, the used “icc” function differentiates between two approaches considering the absolute agreement, which was used in this study if not stated otherwise, and a consistency type. With the latter systematic errors of raters are neglected and only the random residual error is kept.
The agreements were considered with respect to the ICC values poor (<0.5), moderate (0.5–0.75), good (0.75–0.9) or excellent (>0.9) as stated before (21). Plots were generated either using the “ggplot2” (v2.1.0) and the “GGally” (v.1.2.0) packages or by using Microsoft Excel 2013 (Microsoft, Redmond, WA, USA) together with the “Daniel’s XL Toolbox NG” (7.1.4) add-in (www.xltoolbox.net).
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Tissues were used in accordance with the ethical regulations of the NCT tissue bank and approved by the ethics committee of Heidelberg University (S-145/2017).
Results
Tumor cell quantification & inter-rater reliability
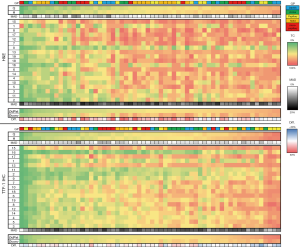
The estimated TCC of all H&E and TTF-1 stained images assessed with the two digital pathology applications (Q, H) and by conventional manual evaluation (1–19: participants) are visualized as heat map in Figure 2. The mean absolute deviation (MAD), as an indicator of variability, was significantly smaller (both P<0.01) between the two software solutions for both stainings compared to inter-rater variability of the conventional estimators, with average MAD values of 3.4 and 2.6 compared to 12.4 and 11.2, respectively. Further, differences between the average digital (ØdPat) and conventional (Øconv) TCC estimates for each case were significantly (P<0.01) smaller when using TTF-1 IHC sections.
In order to evaluate the inter-rater reliability of the TCC estimations, we calculated ICC (ICC using the more stringent “agreement type”, considering absolute differences and therefore also systematic errors) for different groups and both stainings separately. First, considering the reliability within the estimator groups (digital pathology and conventional estimators, comprising BP, PT and NP; Figure 3A) only a poor correlation with values of 0.48 (H&E) and 0.47 (TTF-1), respectively, was observed for the conventional evaluation. In contrast, the concordance between the two digital approaches was high for both stainings, with a good (0.87) and excellent correlation (0.93) pointing towards an improvement in performance using TTF-1 IHC. Further, subgrouping of the conventional estimations according to the level of pathological experience did not reveal a significant difference between the three groups with similar ICC values and highly overlapping confidence intervals.
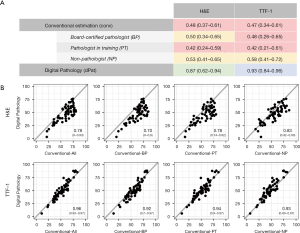
Next, we determined the agreement between the digital and conventional evaluations by calculating ICCs considering the average TCC estimation of each image for the respective approach (Figure 3B). Overall, for H&E stainings a good agreement (0.78) and for TTF-1 IHC an excellent agreement (0.96) was achieved. Looking at the CI intervals and also considering the higher MAD values, we noted that H&E based estimates had a much higher variability in levels of agreement ranging from very poor to excellent (CI: 0–0.93), compared to the TTF-1 based estimation (CI: 0.93–0.97) when data of individual images were considered. The scatter plots show (Figure 3B), that the conventional estimations of TCC based on H&E stainings were consistently higher than the corresponding digital evaluations, as already implied in Figure 2. Additionally, we observed a significant correlation (r=0.81, P<0.01) between the two approaches. Both findings taken together indicated a systematic error (i.e., overestimation of TCC) for conventional H&E based determination of TCC. Indeed, calculation of the ICC using the “consistency type”, which neglects systematic errors (Table S1), yielded an ICC value of 0.89 with a confidence interval ranging from 0.82 to 0.94. Of note, this systematic error was not seen for TCC estimations based on TTF-1 staining.
A corresponding analysis for subgroups with regard to the diagnostic histopathological experience of the estimators yielded comparable ICC values for all subgroups and indicated similar systematic overestimations for H&E stainings. ICC “consistency” values disregarding a systematic error had only minimal variation between the subgroups: BP 0.88 (0.80–0.93), PT 0.87 (0.78–0.92) and NP 0.89 (0.81–0.93).
Influencing factors: histological growth patterns
Next, we sought to identify major factors that may have an impact on the TCC evaluation and could therefore be accountable for higher discrepancies. First, we considered the TCC estimations with regard to the histological growth pattern (Figure S1) and calculated the ICC separately for respective sample subsets (Figure 4A). The resulting agreements were poor/moderate between the conventional estimations and good/excellent between the two digital approaches, comparable to the results for the complete sample set. The ICC values based on the average of the conventional and digital estimations were similar as compared to the total cohort, except for the considerably lower ICC (0.65) regarding H&E stained sections of predominantly solid ADC. A pairwise comparison showed that the TCC was estimated significantly (P<0.01) higher with the conventional approach compared to the digitally supported work-up (Figure 4B). A representative case, HE-S5, exemplifies this finding: the Øconv TCC content of section HE-S5 was estimated to be 70% while the actual TCC was 53% as assessed by dPat (Figure 4C).

Of note, while the confidence intervals of acinar, solid, papillary and micropapillary subsets were in a comparable range as seen for the complete data set, H&E sections of predominant lepidic tumors showed a smaller range of confidence intervals (0.50–0.96).
Influencing factors: TCC
Secondly, we investigated if the TCC estimations were influenced by the actual TCC (determined as ØdPat) of a sample. To this end, we analyzed how many over- or underestimations, defined as a respective difference of 10% to the ØdPat TCC, were carried out following the conventional approach for tumors with a ØdPat TCC of <30%, <40%, 40–60%, >60% and >70% (Figure 4D). Regarding H&E stainings, a general trend for overestimations (39–67%), was observed for tumors with a ØdPat TCC <70%. However, we also noted that the frequency of underestimations increased the higher the ØdPat TCC of a sample was. The proportion of overestimation was significantly (P<0.01) reduced when TCC estimations were based on TTF-1 IHC, both for the entire cohort as well as in matched comparisons for the different TCC subgroups. For both stainings, tumors with a ØdPat TCC between 40% and 60% had the lowest ratio of estimations (H&E: 24%, TTF-1: 45%) within the 10% difference. Of note, almost no underestimations were seen for H&E stainings with a TCC below 30% and almost no overestimations for TTF-1 IHC with a TCC higher than 70%.
Intraobserver reliability of TCC estimations
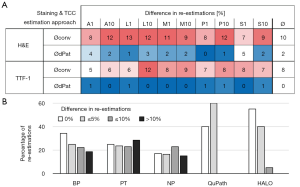
Lastly, we evaluated the consistency of the TCC estimations. To this end, we selected 20 images (two images per growth pattern for both stainings) out of the already evaluated 100 images and rotated them by 180° (Figure 5). The difference of the TCC estimation of the original to the flipped image was calculated to determine the consistency. For the conventional approach the average difference between both estimations was 10.1% for H&E stainings, 7.6% for TTF-1 stainings and 8.5% combined. The digital evaluation yielded significantly lower differences (P<0.01 for all three paired tests) with an overall difference of 1.35%, as well as 2.2% and 0.5% for H&E and TTF-1 staining, respectively. Also, the average differences of estimations based on TTF-1 staining were significantly smaller (conv: P=0.02, dPat: P<0.01) compared to H&E derived estimations for both approaches. Considering the different subgroups of conventional evaluators, the highest consistency was seen for the BP, with no difference between both estimations in 35% of the cases. Only the digital assessments yielded a higher proportion of identical TCC estimations (QuPath: 40% and HALO: 55%).
Discussion
In the present study, we investigated the variability of manual conventional and semi-automatic digital quantification of the TCC in ADC and determined aspects improving or impairing reliable estimations. We found that (I) the agreement of TCC estimation between the two digital software solutions was higher compared to the agreement within the group of 19 participants, (II) that the agreement of the average digital TCC estimation to the average human TCC estimations was good, (III) that the agreement was improved using TTF-1 immunohistochemical staining, (IV) and that the agreement was significantly reduced for the conventional estimation of H&E sections of tumors with a solid growth pattern. Within the different subgroups of conventional estimators, the average agreement was comparable regardless of the level of experience. Moreover, it became clear that estimators tend to overestimate the TCC in tumors especially for H&E stainings and that the highest rate of misclassification was seen for tumors with about 50% TCC. Finally, we could show that the consistency of estimations was higher for the semi-automatic digital evaluation with only 1% difference in average compared to 9% for the conventional analysis approach. The web application that was developed and used for this study including the complete image set is freely available for self-testing to interested parties (www.hd-molpath.de/tcc-test) and also features an additional training mode (www.hd-molpath.de/tcc-trainer).
Dufraing et al. have already anticipated the potential of computer-assisted TCC estimations when they hypothesized that digital tools, thoroughly validated and trained to recognize neoplastic cells, might be a solution to reduce interobserver variation. It was shown previously that mean estimates from multiple observers are fairly accurate (22). This is in keeping with our results showing a good agreement of computer-aided TCC estimations with the average of human TCC estimations. Therefore, we conclude that digital software solutions are a suitable support system that can reduce interobserver variability (Figure 6).
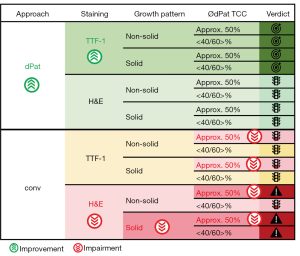
All samples included in our study were TTF-1 positive ADC. As such TTF-1 IHC could be used to highlight the tumor cell nuclei and was expected to simplify TCC estimations and lower interobserver variation. We could demonstrate an improvement of the agreement between the two software tools as well as for the agreement of the averages of computer-aided and conventional evaluations, but unexpectedly the agreement within the human observers remained poor. This finding indicates that basic recognition of tumor cells might not be a major confounding aspect for the conventional TCC quantification. Similar conclusions can be drawn considering the different diagnostic histopathological experience of our participants, as the performance of trained NP was non-inferior to the performance of pathologists. A large survey including 105 laboratories across Europe revealed that in the majority of laboratories TCC is estimated by pathologists (approx. 90%), but in a subset also by molecular biologists or technicians followed by confirmation by a pathologist (22). In this study, no significant difference was observed between these groups. Our study supports that finding although we strongly advise confirmation by an experienced pathologist. Even a slightly better agreement was observed for NP compared with the computer-aided evaluation regarding H&E stainings, which might be explained by the time spend for the analysis which was not measured here.
Other large interobserver studies also showed that TCC estimations are highly variable among pathologists (7-9,22). For example, a large multi-institutional study including 194 participating laboratories revealed that although the concordance of 8 out of 10 images was within 10% of the criterion standard, images with the highest deviation exhibited TCC counts ranging from 10% to 95% (9). Our results are well in line with the literature, as the agreement between observers was poor [ICC: 0.48 (HE) and 0.47 (TTF-1)].
To the best of our knowledge, our study is the first to consider the growth pattern of ADC in the evaluation of TCC. While the agreement within and between the conventional and digital evaluation was similar to the complete data set for predominant lepidic, acinar, papillary or micropapillary growth patterns, only a moderate agreement was seen for H&E stained ADC with solid growth pattern. The significant overestimation seen by the conventional evaluators might be due to the fact that tumor cells within a solid growth pattern are considered less differentiated and generally larger creating the impression of an overall larger tumor region. It seems visually more difficult to assess TCC when cells of different sizes (tumor, stromal and immune cells) need to be considered separately.
Further, we perceived that conventional TCC estimation was influenced by the genuine TCC of a sample. The highest ratio of misclassification (±10%) was seen for samples with an actual TCC around 50% for both stainings (H&E: 76%, TTF-1: 55%) highlighting an additional caveat for tumor samples in this range. For H&E stainings, overestimation was eminent in the majority of cases, as reported before (7-9,22). This can be critical for samples with a low TCC leading to false-negative or incomplete sequencing reports and therefore harbors a significant potential for diagnostic errors (23). Smits et al. reported in samples with a TCC below 20% an overestimation in 38% of cases (8), which concurs with our observation of an overestimation in 47% of samples with a TCC below 30%. To reduce this rate, we recommend the use of TTF-1 IHC (that is often already performed as part of the diagnostic routine workup) and/or digital evaluation for low TCC samples. Although not analyzed by us, it is reasonable to assume that the use of other commonly used IHC markers (CK7, Napsin) can also support TCC measurement, especially for TTF-1 negative tumors. Future studies might help clarifying this issue.
Finally, we assessed the intraobserver variability considering 180° rotated images. Consistency of the software was expectedly very high, but not perfect. Apparently, the different arrangement of the input pixels resulted in minor changes during the cell recognition steps which might lead to the minor differences (1% on average). However, digital approaches still exceeded the consistency of the conventional approach (9% on average), which further demonstrates the potential of improvement of the TCC estimation.
The image size of 1.5–2 mm2 used for TCC evaluation in this study, was selected in order to simulate the tissue size of biopsy specimens, which make up the great majority of NSCLC samples used for molecular diagnostics. Further, this image size enabled the evaluation of the complete image at a glance, ensuring that evaluations were not biased by the way the images were inspected (scrolling, focus on specific areas). However, an interesting aspect for a subsequent study would be to investigate the concordance of TCC estimates in whole slide images.
In summary, our results show several aspects improving or impairing reliable TCC estimations. Considering conventional vs. digital estimations, H&E and TTF-1 staining, the histological growth pattern, the actual TCC and the consistency of estimations we see a high potential for semi-automatic digital TCC estimation, particularly prior to costly and time-consuming molecular diagnostic workflows. With the increasing availability of affordable whole-slide imaging systems and a variety of commercial as well as open source software solutions a broader introduction into clinical practice would be possible within the near future. Further we provide a free web application comprising all images of this study and corresponding ØdPat TCC as reference for training (www.hd-molpath.de/tcc-trainer) and self-testing (www.hd-molpath.de/tcc-test).
Acknowledgments
We thank Veronika Geißler and Terence Osere for excellent technical assistance, as well as the Tissue Bank of the National Center for Tumor Diseases, Heidelberg. Germany for expert handling of the tissue.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-1168
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-1168
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-1168). DK reports personal fees from AstraZeneca, Bristol-Myers Squibb, and Pfizer outside the submitted work. ALV reports personal fees from Astra Zeneca, outside the submitted work. RL reports grants from Deutsche Forschungsgemeinschaft (DFG), Dietmar Hopp Stiftung, outside the submitted work. CPH reports grants from Siemens, Pfizer, MeVis, Boehringer Ingelheim, German Center for Lung Research, personal fees from Astellas, AstraZeneca, Basilea, Bayer, Boehringer Ingelheim, Bracco MEDA Pharma, Chiesi, Covidien, Essex, Fresenius, Gilead, Grifols, Intermune, Lilly, MSD, Novartis, Pfizer, Pierre Fabre, Roche, Schering-Plough, Siemens, other from GSK, outside the submitted work; in addition, CPH has a patent Method and Device For Representing the Microstructure of the Lungs, IPC8 Class: AA61B5055FI, PAN: 20080208038 issued. FH reports personal fees from Uptake, BTG, Olympus, Pulmonx, outside the submitted work. PC reports grants and personal fees from Novartis, Roche, AstraZeneca, Takeda, and personal fees from Pfizer, Chugai, Boehringer, outside the submitted work. TM reports grants and non-financial support from Roche Diagnostics GmbH, Penzberg, Germany, outside the submitted work; in addition, TM has a patent WO2019158460 pending, a patent WO2019211418 pending, a patent WO2019215223 pending, a patent EP3391053 issued, and a patent EP3365679 pending. MR reports personal fees from Amgen, AstraZeneca, Boehringer-Ingelheim, BMS, Lilly, Celgene, Merck, Mirati, MSD, Novartis, Pfizer, Roche, Samsung Bioepis, outside the submitted work. WW reports personal fees from Roche, MSD, BMS, AstraZeneca, Pfizer, Merck, Lilly, Boehringer, Novartis, Takeda, Amgen, Astellas, Illumina, Agilent, Siemens, Molecular Health and grants from Roche, MSD, BMS, Bruker, AstraZeneca, outside the submitted work. JB reports grants from German Cancer Aid, outside the submitted work. MT reports grants, personal fees and non-financial support from AstraZeneca, Bristol-Myers Squibb, Takeda, Roche, personal fees and non-financial support from AbbVie, Boehringer Ingelheim, Celgene, Chugai, Lilly, Novartis, Pfizer, outside the submitted work. SP reports personal fees from Abbvie, Amgen, AstraZeneca, Bayer, Biocartis, Boehringer-Ingelheim, BMS, Clovis, Daiichi Sankyo, Debiopharm, Eli Lilly, F. Hoffmann-La Roche, Foundation Medicine, Illumina, Janssen, Merck Sharp and Dohme, Merck Serono, Merrimack, Novartis, Pharma Mar, Pfizer, Regeneron, Sanofi, Seattle Genetics and Takeda, Takeda, Bioinvent, Medscape, Phosphoplatin Therapeutics; non-financial support from Amgen, AstraZeneca, Boehringer-Ingelheim, BMS, Clovis, F. Hoffmann-La Roche, Illumina, Merck Sharp and Dohme, Merck Serono, Novartis, Pfizer, Phosphoplatin; outside the submitted work; all fees to Institution. PS reports personal fees from BMS, MSD, Incyte, Janssen, Amgen, Novartis, Roche and AstraZeneca outside the submitted work. AS reports grants and personal fees from Bayer, BMS, grants from Chugai and personal fees from Astra Zeneca, MSD, Takeda, Seattle Genetics, Novartis, Illumina, Thermo Fisher, Eli Lily, Takeda, outside the submitted work. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Tissues were used in accordance with the ethical regulations of the NCT tissue bank and approved by the ethics committee of Heidelberg University (S-145/2017).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Mosele F, Remon J, Mateo J, et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol 2020;31:1491-505. [Crossref] [PubMed]
- Arbour KC, Riely GJ. Systemic therapy for locally advanced and metastatic non-small cell lung cancer: a review. JAMA 2019;322:764-74. [Crossref] [PubMed]
- Dufraing K, van Krieken JH, De Hertogh G, et al. Neoplastic cell percentage estimation in tissue samples for molecular oncology: recommendations from a modified Delphi study. Histopathology 2019;75:312-9. [Crossref] [PubMed]
- Budczies J, Kazdal D, Allgauer M, et al. Quantifying potential confounders of panel-based tumor mutational burden (TMB) measurement. Lung Cancer 2020;142:114-9. [Crossref] [PubMed]
- Kazdal D, Endris V, Allgäuer M, et al. Spatial and temporal heterogeneity of panel-based tumor mutational burden in pulmonary adenocarcinoma: separating biology from technical artifacts. J Thorac Oncol 2019;14:1935-47. [Crossref] [PubMed]
- Stenzinger A, Endris V, Budczies J, et al. Harmonization and standardization of panel-based tumor mutational burden measurement: real-world results and recommendations of the quality in pathology Study. J Thorac Oncol 2020;15:1177-89. [Crossref] [PubMed]
- Lhermitte B, Egele C, Weingertner N, et al. Adequately defining tumor cell proportion in tissue samples for molecular testing improves interobserver reproducibility of its assessment. Virchows Arch 2017;470:21-7. [Crossref] [PubMed]
- Smits AJ, Kummer JA, de Bruin PC, et al. The estimation of tumor cell percentage for molecular testing by pathologists is not accurate. Mod Pathol 2014;27:168-74. [Crossref] [PubMed]
- Viray H, Li K, Long TA, et al. A prospective, multi-institutional diagnostic trial to determine pathologist accuracy in estimation of percentage of malignant cells. Arch Pathol Lab Med 2013;137:1545-9. [Crossref] [PubMed]
- Dijkstra JR, Heideman DA, Meijer GA, et al. KRAS mutation analysis on low percentage of colon cancer cells: the importance of quality assurance. Virchows Arch 2013;462:39-46. [Crossref] [PubMed]
- Mukhopadhyay S, Feldman MD, Abels E, et al. Whole slide imaging versus microscopy for primary diagnosis in surgical pathology: a multicenter blinded randomized noninferiority study of 1992 cases (pivotal study). Am J Surg Pathol 2018;42:39-52. [Crossref] [PubMed]
- Pell R, Oien K, Robinson M, et al. The use of digital pathology and image analysis in clinical trials. J Pathol Clin Res 2019;5:81-90. [Crossref] [PubMed]
- Bankhead P, Loughrey MB, Fernandez JA, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep 2017;7:16878. [Crossref] [PubMed]
- Aeffner F, Zarella MD, Buchbinder N, et al. Introduction to digital image analysis in whole-slide imaging: a white paper from the Digital Pathology Association. J Pathol Inform 2019;10:9. [Crossref] [PubMed]
- Huss R, Coupland SE. Software-assisted decision support in digital histopathology. J Pathol 2020;250:685-92. [Crossref] [PubMed]
- Grabe N, Roth W, Foersch S. Digital pathology in immuno-oncology-current opportunities and challenges: overview of the analysis of immune cell infiltrates using whole slide imaging. Pathologe 2018;39:539-45. [Crossref] [PubMed]
- Warth A, Muley T, Meister M, et al. The novel histologic International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification system of lung adenocarcinoma is a stage-independent predictor of survival. J Clin Oncol 2012;30:1438-46. [Crossref] [PubMed]
- Horai Y, Mizukawa M, Nishina H, et al. Quantification of histopathological findings using a novel image analysis platform. J Toxicol Pathol 2019;32:319-27. [Crossref] [PubMed]
- Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol 2015;10:1243-60. [Crossref] [PubMed]
- Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol Bull 1979;86:420-8. [Crossref] [PubMed]
- Koo TK, Li MY. A Guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med 2016;15:155-63. [Crossref] [PubMed]
- Dufraing K, De Hertogh G, Tack V, et al. External quality assessment identifies training needs to determine the neoplastic cell content for biomarker testing. J Mol Diagn 2018;20:455-64. [Crossref] [PubMed]
- Endris V, Penzel R, Warth A, et al. Molecular diagnostic profiling of lung cancer specimens with a semiconductor-based massive parallel sequencing approach: feasibility, costs, and performance compared with conventional sequencing. J Mol Diagn 2013;15:765-75. [Crossref] [PubMed]


