
Deep sequencing reveals the genomic characteristics of lung adenocarcinoma presenting as ground-glass nodules (GGNs)
Introduction
Lung cancer is a leading cause of cancer-related deaths worldwide (1). Currently, the majority of lung cancer cases are diagnosed at advanced stage, and despite improvements in molecular diagnosis and targeted therapies, the average 5-year survival rate for lung cancer remains less than 20% (2). Limiting these advances is a poor knowledge of the earliest events that underlie lung cancer development and that would constitute markers and targets for early detection and prevention. Understanding the genomic variations of early stage lung cancer may reflect the initial features of tumorigenesis, and is crucial for the proper management of lung cancer.
Ground-glass nodules (GGNs) are characterized as nodules with ground-glass opacity (GGO) in lung parenchyma, which has been described as haziness with increased lung attenuation by computed tomography (CT) and preserved bronchial and vascular margins (3-5). Several studies have shown that persistent GGN on CT is associated with lung adenocarcinoma (LUAD), which should be suspected with a high risk of malignancy (6,7). With recent advances in diagnostic imaging modalities and the widespread use of chest CT screening, the detection rate of lung ADC presenting as GGNs is increasing. GGNs generally grow slowly, have a good prognosis (8), and are considered an early stage of tumorigenesis (9,10).
Accumulating studies have analyzed the characteristics of GGNs in various aspects, including radiology, pathology, surgery, and molecular biology, providing information on emerging and rapidly progressing aspects of surgical treatments for GGNs (11-14). However, the distinct genomic profiles involved in GGN progression and their potential for guiding therapeutic strategies have not yet been defined. According to different clinical characteristics, GGNs could be divided into different categories. GGNs are radiologically divided into the following two categories: pure GGNs, which contain no solid components, and mixed GGNs, which contain both a pure GGO region and a consolidated region (15). Moreover, GGNs are generally divided into different categories according to the number, i.e., solitary or multiple, as well as the lepidic components, i.e., lepidic or invasive ADC (16). Whether genetic alterations are associated with the clinical characteristics remain unsolved.
Here, we presented the first comprehensive description of the genomic architecture of GGNs by whole-exome deep sequencing (>1,000×) and amplicon deep sequencing (~30,000×). The comprehensive analysis, including genetic alterations, intratumor heterogeneity (ITH), frequent events, and mutational signatures, was performed for the samples. The genetic alterations associated with clinical characteristics of GGNs were analyzed.
We present the study in accordance with the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-1086.
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Human Research Ethics Committee of Beijing Cancer Hospital and Beijing Institute of Genomics (2015KT71), and informed consent was obtained from the patients.
Patient and samples
Consecutive patients who had been diagnosed with primary lung cancer and underwent surgical resection in Peking University Cancer Hospital (Beijing, China) between 2012 and 2017 were recruited for this retrospective study. Of these, 28 patients were selected based on having GGN in the preoperative CT examination. Primary tumors at stage IIA or greater and tumors without GGN features in the CT examination were excluded. These patients were not treated with neoadjuvant chemotherapy or radiation therapy, and all the nodules from these patients were diagnosed as early-stage LUAD (stage I). Exclusion criteria included those cases did not meet the criteria of containing at least 20% of tumor cells by pathologists, as well as those without the adjacent normal tissue specimens. This series included 30 lesions detected from 28 patients, and two of them contributed two samples respectively (P17, R31, see Table S1).
Clinical features
Histopathologic diagnoses of GGNs were according to the new IASLC/ATS/ERS multi-disciplinary. Lepidic-predominant adenocarcinomas (LEPs) and non-LEPs with predominant invasive components, such as acinar, papillary, and micropapillary ADC were defined (Table S1 and Figure S1).
CT images were interpreted by two experienced thoracic radiologists in Beijing cancer hospital. GGN is defined as hazy increased attenuation of the lung tissue with preservation of the bronchovascular structures. The lesions were classified as (I) pure GGO (pGGO), no solid part of the nodule; (II) mixed GGO (mGGO), GGO with a solid part occupying less than 50% of the nodule (Table S1 and Figure S1).
Clinical information including gender, age at diagnosis, smoking status, pathological TNM stage, tumor location, lymphatic invasion, visceral pleural invasion were collected for further analysis.
Whole-exome sequencing (WES)
Genomic DNA was extracted from the tumor and adjacent normal tissues using the QIAamp DNA Mini Kit (Qiagen 51304). Purified 100 ng–1 µg genomic DNA from each sample was sonicated using Covaris S220. Libraries were constructed from each sample with the Agilent SureSelectXT2 (Illumina) according to the manufacturer’s instructions and were further captured using the Agilent SureSelect Target Enrichment System (Human All Exon V6 Kit). Paired-end sequencing of 2×150 bp was performed on the Illumina HiSeq X Ten platform at Novogene. The sequencing depths of each sample are listed in Table S2.
WES data processing
Paired-end data were aligned to the human reference sequence (UCSC hg19) using the Burrows-Wheeler Aligner (17,18). All the aligned reads were further processed using Picard tools and Genome Analysis Toolkit (GATK) (19) and included deduplication, base quality recalibration, and multiple-sequence realignment prior to mutation detection.
Identification of single-nucleotide variations (SNVs) and indels
To identify somatic variations, adjacent normal tissues were used as the normal control. Somatic SNVs were identified using MuTect (19). The passed variants were further filtered using the following described criteria to obtain a more confident set of SNVs: (I) SNVs in the corresponding normal sample were filtered out; (II) the SNVs showing a mutation frequency of more than 10% in a tumor sample (double-strand support) were maintained; (III) the SNVs with a frequency of less than 10% in the tumor were filtered by Shearwater (20), an algorithm to detect high-confident variants at a low frequency. Only variants that were significantly mutated over the error model were kept, using a q value cutoff of 0.05 by multiple testing; and (IV) the dbSNP germline mutations (dbSNP138 version) were filtered out from the SNV list.
VarScan2 (21) was employed to investigate somatic indels. The indels showing more than four variant reads (double-strand support) in a tumor sample and none in the corresponding normal tissue sample were kept for further analysis. Both somatic SNVs and indels were subsequently annotated by multiple databases using the ANNOVAR tool (22).
Targeted amplicon deep sequencing
We performed targeted amplicon deep sequencing to validate SNV calling from the WES data. We randomly selected 130 SNVs calling from p9, p11, p14 and p17t2. Multiple PCR primers for amplicons containing 130 target SNVs were designed by Ion AmpliSeq Designer and implemented using the Ion AmpliSeq™ Library Kit 2.0. Targeted amplifications were used to construct the next-generation sequencing libraries. The libraries were constructed using the NEBNext® Ultra™ End Repair/dA-Tailing Module and the NEBNext® Ultra™ Ligation Module according to the manufacturer’s instructions. The ligation productions were purified by AMPure XP Beads and amplified by KAPA HiFi HotStart ReadyMix. The final sequencing libraries were obtained after PCR purifications. Subsequently, paired-end sequencing of 2×150 bp was performed on the Illumina HiSeq X Ten platform to obtain a depth of ~30,000×.
Variants calling from targeted amplicon deep sequencing
Sequence reads were mapped against the human reference genome hg19 using BWA (17). The bam files were realigned and recalibrated using GATK (19). Samtools mpileup (23) were used for extracting the frequency of selected SNVs from both tumor and normal tissue samples.
Somatic copy number analysis
We used Sequenza software (24) to estimate CNAs, cellularity and ploidy in GGNs. We determined genomic instability by using the methods according to Nahar et al. (25). In brief, the genomic instability index (GII) was calculated as the fraction of the total genome which was altered with a copy change ≥1 relative to the median integer ploidy. The amplification and deletion-based genomic instability index (adGII) was defined as the fraction of the total genome affected by high-copy gains and losses (or amplification and deletions with copy change ≥2 relative to ploidy).
Determination of genome-doubling status
The genome-doubling status for each GGN was estimated by a previously published algorithm (26). In brief, the P value was obtained using 10,000 simulations with observed probabilities of copy-number events. For samples with a ploidy ≤3, a P value threshold of 0.001 was used. To avoid underestimating genome doubling in high-ploidy samples, a P value threshold of 0.05 was used for samples with a ploidy =4, and all samples were classified as being genome doubled if the ploidy exceeded 4.
Determination of the cancer cell fraction (CCF) and timing of mutations
The CCF and mutant allele copy number for a given SNV were calculated by an algorithm previously described (27). Mutations were classified as “clonal” if the 95% confidence interval of CCF exceeded 1; otherwise, mutations were classified as “subclonal”.
As previously described (27), the timing of mutations relative to copy-number alteration or WGD was defined on the integer mutant allele copy-number. Briefly, in samples with WGD, mutations were classified as “pre-WGD” when the integer mutant allele copy number was ≥2, while any mutations with a mutation copy number of 1 were classified as “post-WGD”.
Mutation signature analysis
All the SNVs were categorized into six types, including C > A, C > G, C > T, T > A, T > C, and T > G. Classification of the substitutions was further refined by including flanking 5’ and 3’ bases of each mutated site. For example, T > A could be characterized as ATG > AAG when the 5' site was an A and the 3' site was a G. Considering all the possibility in the 5' and 3' bases, there would be 96 types of substitutions. Subsequently, the profiles of 96 tri-nucleotide mutational contexts for each sample were used for detecting the mutational signatures of GGNs by the R package “Mutational Patterns” (28). The correlation coefficients were calculated between the estimated signatures and the known COSMIC signatures. The known signatures showing the maximum cosine similarity were defined as the mutational signatures in GGNs.
Driver genes and regions analysis
We defined potential LUAD driver genes (n=78) as previously described (25). Significantly mutated genes in GGNs were identified using both MutSigCV2.0 (29) and dNdScv algorithms (30) with a q value cutoff of 0.1. The significance of broad and focal CNAs was assessed from the segmented data using the GISTIC 2.0 algorithm (31). We performed functional enrichment for the genes located in the recurrent CNA regions using DAVID (32).
Neoantigen prediction
POLYSOLVER (33) was employed for HLA typing. We used nonsynonymous mutations to generate a list of peptides ranging from 9–11 amino acids in length with the mutated residues. Predictions for the binding affinity of every mutant peptide and its corresponding wild-type peptide to the patient’s germline HLA alleles were performed using the NetMHCpan 4.0 algorithm (34). Candidate neoantigens were identified as those with a predicted strong or weak mutant peptide binding affinity and no binding affinity of its corresponding wild-type peptide. The clonality of neoantigens was defined on the clonal status of the corresponding mutations.
Statistical analysis
Pearson correlation coefficients were used to evaluate the association between mutated allele frequency calling from the WES and the amplicon deep sequencing, as well as the neoantigen burden and exonic mutation burden among GGNs. R2 was used to depict the squared Pearson correlation coefficient. For comparisons of pathological subtypes between genotypes, P values were based on the Wilcoxon test for categorical variables and two-sample t-test for continuous variables.
Results
Histological and radiological examination
We retrospectively collected 30 lung cancer samples from 28 patients presenting as GGNs on computed tomography (CT) scans. All patients underwent GGN surgical resection and were determined to have early-stage LUAD (stage I). These GGNs were classified into 8 pure GGNs and 22 part-solid nodules (mixed GGNs) based on chest CT results. According to the histological subtype of adenocarcinoma present in the sample, these GGNs were divided into two groups as follows: 6 lepidic-predominant GGNs, as LEPs, and 24 non-LEPs with predominant invasive components, such as acinar, papillary, and micropapillary adenocarcinoma. Comprehensive clinical information and images are provided in Table S1 and Figure S1. We examined the potential relationship between iconography and histologic patterns. Notably, the mixed GGNs tended to accompany a pathologically nonlepidic growth pattern (Fisher’s exact test, P<0.05), indicating a high correlation between radiological and histological examinations. This finding was consistent with previous observations in LUAD (35,36), which reported an association between histological subtypes and GGO features. Due to the low tumor purity shown in pathological detection (Table S1), deep sequencing must be carried out in order to achieve high sensitivity and accuracy in mutation calling.
Deep WES
WES was performed for 30 surgically resected GGNs from 28 patients (two of the patients with multifocal nodules) to obtain a depth of ~1,000× depth for tumor samples and ~300 for adjacent tissue (Table S2). The adjacent tissue in each patient was used as a matched normal control for somatic variation calling (see Methods section). A total of 130 somatic single-nucleotide variants (SNVs, frequency: 0.5–21.4%, Table S3) randomly selected from p9, p11, p14 and p17t2 were subjected to targeted deep sequencing (mean depth of 30,000×). The results validated by target deep sequencing were highly correlated with the results from WES (R2=0.964, Pearson’s correlation coefficients, Figure S2), confirming the reliability of the SNV calling. In total, we identified 4,230 SNVs and 340 indels in GGNs (see Methods section, Tables S4,S5 and Figure 1A). The average number of somatic variations among the samples was 152 (range, 77–429 variations), corresponding to a median of 2.33 variations/MB and a mean of 2.54 variations/MB (range: 1.28–7.15 variations), showing a relatively low mutational burden in GGNs compared with the results of the TCGA LUAD sequencing study (37), but quite close to the burden in nonsmoking LUADs (25,38).
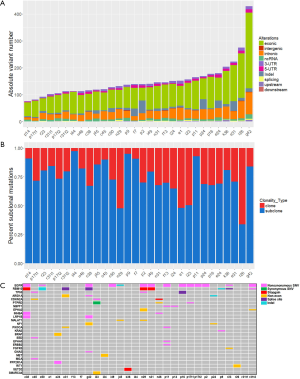
The clonality of functionally significant somatic mutations
We estimated the CCF (see Methods section) of the mutations for each sample, and identified 1,243 (27.2%) clonal mutations and 3,327 (72.8%) subclonal mutations (Tables S4,S5 and Figure 1B). The percentage of subclonal mutations was employed to determine ITH, and a mean of 74.2% ITH (range, 33.8–97.3%) in GGNs was observed, which was higher than previous findings of ITH in lung cancer sequencing studies (~30% branch mutations) (39-41), but comparable to findings of the EGFR-mutant LUADs (25).
We surveyed known cancer genes (see Method section) for potential driver mutations and found that 25 samples (83.3%) contained at least one variant in a gene known to be involved in LUAD (Figure 1C). Alterations of 18 LUAD-associated genes in certain samples were found to be subclonal, including targetable mutations in EGFR (Figure 1C). This result informed potential therapeutic strategies, since target subclonal alterations that present in only a proportion of cells may result in reduced treatment efficacy. We then applied both MutSigCV and dNdScv tools to identify significantly mutated genes (SMGs) with statistically higher-than-expected mutation prevalence across the entire patients (q <0.1). Both methods identified only two SMGs: EGFR (frequency: 46.7%, 14/30 samples; 11 clonal/3 subclonal; 4 male/9 females) and RBM10 (frequency: 30.0%, 9/30 samples; 6 clonal/3 subclonal; 1 male/8 females). Epidermal growth factor receptor (EGFR) is the most common therapeutically targetable driver for LUADs. In these dataset, 11 samples harbored EGFR L858R mutation (10 clonal/1 subclonal), which was a recurrent activating mutation within the EGFR kinase domain (42). RBM10 encodes an RNA-binding protein, and is subject to recurrent inactivating mutations in LUADs (43). Notably, the GGNs with RBM10 mutations tended to accompany a pathologically lepidic pattern (Fisher’s exact test, P<0.05), indicating better outcomes of patients with lepidic tumors than nonlepidic tumors.
Mutational signatures during progression of GGNs
The distinguished clonal (trunk) and subclonal (44) mutations in each sample were further used to detect the mutational signatures in GGNs. The distributions of identified mutational signatures in clonal and subclonal mutations were heterogeneous among the patients, and were also heterogeneous in different nodules within the same patient, i.e., p17t1 and p17t2 (Figure 2A).
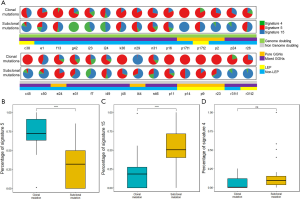
Among the three signatures identified in our cohorts, molecular clock-like signature 5 possessed a significantly higher proportion in clonal mutations (Figure 2B, P value =1.6E-07), while defective DNA mismatch repair-associated signature 15 became more dominant in subclonal mutations (Figure 2C, P value =2E-06). In signature 4, no significant differences were observed between trunk and branch mutations (Figure 2D, P value =0.09). These results revealed that different mutational processes were operative during the progression of GGNs. Notably, an incongruous mutational signature pattern was observed in one patient, l44, in which signature 15 dominantly contributed to the mutational landscape and tended to be more dominate in clonal mutations than in subclonal mutations (Figure 2A).
Copy number alterations in GGNs
We identified a total of 7001 CNAs at a median of 216 CNAs per sample (Figure S3), which is fewer than in the previous large-scale lung cancer study (41). We also examined the known recurrent copy number alterations in LUADs as previous described (25), and almost all the known gains and deletions were observed in at least one sample (Figure S4). We assessed a GII (defined as fraction of the genome altered by CNAs, copy change ≥1 relative to ploidy; see Methods section), and observed that the majority of tumors showed low-to-moderate genomic instability (median of 19.6% per tumor, Figure 3A). A median of only 1.8% of the genome was affected by high-copy gains and losses (copy change ≥2 relative to ploidy; defined as adGII); see Methods section; Figure 3B). Among all the GGNs, we found that non-LEPs harbored significantly higher GII scores than LEPs (Figure 3C, P=0.041, Wilcoxon test), indicating a higher degree of malignancy in non-LEPs (45,46). Increased GII scores were also detected in the mixed GGNs compared to pure GGNs, although these differences were not statistically significant (Figure 3D, P=0.5, Wilcoxon test).
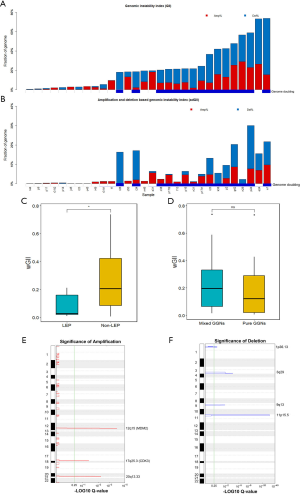
GISTIC 2.0 analysis (with a threshold of q <0.25) revealed 3 focal amplifications and 4 focal deletions recurrently altered in GGNs along with 14 recurrently altered whole arms (Figure 3E,F and Table S6). Among the recurrent focal regions, 12q15 (23.30%), 17q25.3 (50.00%), and 20q13.33 (36.70%) amplifications and 1p36.13 (66.70%) and 11p15.5 (70.00%) deletions have been previously found in LUAD (37,47-49). Deletion events occurring at 11p15.5 and 3q29 were previously reported to be associated with asbestos-related lung cancer and lung cancer susceptibility in Koreans, respectively (50,51). Notably, 12q15 encodes an oncogene, MDM2, which has been reported as a frequently amplified gene in LUAD (43). In addition to previously reported regions, two novel recurrent deletions were identified at 3q29 (43.30%) and 9q13 (66.70%).
To detect the potential functional effect of recurrently focal events, we performed Gene Ontology (GO) analysis for the genes located in the amplified and deleted regions (Table S7), respectively. Among the significant terms of amplified genes (Table S8), regulation of cell proliferation, regulation of growth and negative regulation of apoptotic process were highly associated with tumorigenesis. On the other hand, the deleted genes were enriched in some metabolic process, i.e., arachidonic acid secretion and lipid catabolic process (Table S8). These results highlight the potential functional roles of recurrent CNAs in the formation of GGNs.
Early emergence of genome doubling in GGNs
In total, 90% of the samples were estimated as aneuploidy, and the ploidy ranged from 1.5 to 6.6 (Table S9). We then applied an algorithm to identify tumors that were likely to have undergone a genome-doubling event, even if they are no longer polyploidy (see Methods section). Among all the 30 samples, 15 GGNs (50%) showed genome- doubling status (Table S9), indicating that whole-genome doubling (WGD) is a frequent event in GGNs. The inferred CCFs and timing of the mutations relative to WGD showed that 10.7–67.1% clonal mutations occurred after the WGD (defined as post-WGD mutations, see Methods section; Figure 4A), indicating that WGD was a truncal event wherever present and occurred during the tumorigenesis.
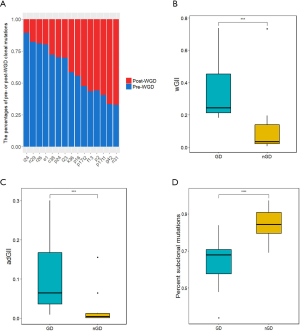
Significantly higher GII and adGII scores were observed in genome-doubled samples compared to nondoubled samples (Wilcoxon test, P value <0.05, Figure 4B,C), indicating that WGD was associated with significantly higher genomic instability. This result was in agreement with the accumulating evidence showing that genome-doubling events are associated with the propagation of genome instability (26,52,53). There is no evidence that WGD was associated with clinical characteristics (solid portions, Fisher’s exact test, P=0.94; non-Lepidic growth pattern, Fisher’s exact test, P=0.073), RBM10 mutation (Fisher’s exact test, P=1), and EGFR mutation (Fisher’s exact test, P=0.27). However, there were significant associations between WGD events and mutational ITH, as the proportion of subclonal mutations was significantly lower in the WGD samples than in the non-WGD samples (Figure 4D, P=9.7E-05, Wilcoxon test), which suggests a relatively longer trunk in the WGD samples.
Association between mutation burden and clinical characteristics in GGNs
To investigate whether there are differences in the mutation burden between GGNs with distinct clinical features, we further classified these samples into different groups based on iconography and histologic patterns (Table S1). We observed that mixed GGNs occupied significantly more exonic mutations than pure GGNs (Figure 5A, P=0.017, Wilcoxon test). Previous observations in clinical cases showed the progression from nonsolid to part-solid nodules (10,54), which was consistent with the fact that more mutations accumulated in mixed GGNs than in pure GGNs. This result may reflect the fact that pure GGNs are found at an earlier stage of carcinogenesis. Likewise, a significantly increased exonic mutation burden was also observed in nonlepidic GGNs compared to lepidic GGNs (Figure 5B, P=0.0064, Wilcoxon test), indicating that a lower mutation burden was highly associated with the lepidic growth pattern. Moreover, when we divided each pure and mixed sample into sub-groups with lepidic or nonlepidic growth patterns, the statistical significance of the differences in the exonic mutation burden was observed between mixed GGNs with non-LEP patterns and pure GGNs with LEP patterns (Figure 5C, P<0.05, Wilcoxon test). Previous studies have shown the positive association between the mutation burden and patient survival in the setting of anti-PD-1 therapy (55,56). This association highlights the potential of immunotherapeutic strategies for a subset of lung cancers.

Some of the mutations could create neoantigens, which are foreign to immune systems and capable of inducing antitumor immune responses. To investigate the neoantigen landscape in GGNs, we predicted neoantigens among the patients (Figure S5). The neoantigen burden was highly associated with the exonic mutation burden (Figure S6), which was consistent with previous studies (57,58). Likewise, we observed significantly higher clonal neoantigen burden in non-LEPs than in LEPs (Figure 5D, P=0.036, Wilcoxon test). We also observed a trend toward higher clonal neoantigen burden in mixed GGNs than in pure GGNs, although the differences between the two groups did not reach statistical significance (Figure 5E, P=0.17, Wilcoxon test). Previous research suggested that neoantigen heterogeneity influences immune surveillance and support therapeutic developments targeting clonal neoantigens (58). This research highlights that there might be more neoantigens to be targetable and effective in a subset of lung cancer.
Common or independent origins among GGNs within patients
Whether multiple GGNs represent as independent origination may influence the treatment and prognosis. We detected the evolutionary relationships between the multifocal GGNs in our dataset. Patient r31 presented with two GGNs, including t1 in right middle lobe and t2 in right upper lobe. The r31t1 and r31t2 displayed as mixed GGNs with different major histological subtypes, presenting as non-LEP and LEP dominant, respectively. These two lesions had no mutations in common, indicating that they are independently originated. Three known LUAD-associated gene, EGFR, RBM10, and EPHA2, were found to be mutated solely in r31t2 (Table S4).
Patient p17 harbored two pure GGNs with different major histological subtypes, presenting as non-LEP and LEP dominant, respectively. The p17t1 (in superior segment of left lower lobe) and p17t2 (in basal segment of left lower lobe) shared 16 exonic mutations (Table S4), including EGFR L858R mutation, indicating that they originated from a common ancestor and that intrapulmonary metastasis has occurred even in early stage of lung cancer, in agreement with the previous report by Li et al. (59). Furthermore, amplification of an oncogene MYCL1 and WGD event were also detected in the two lesions.
Discussion
Through >1,000× WES and ~30,000× amplicon deep sequencing, we have, for the first time, characterized the genomic landscape of GGNs. Despite the relatively low somatic mutation burden (a median of 2.33 variations/MB and a mean of 2.54 variations/MB) compared to the TCGA LUAD sequencing study (37), GGNs exhibited high ITH characterized by the proportion of subclonal mutations (range, 33.8–97.3%) in each sample. Subclonal mutations were found in certain LUAD-associated genes, i.e., the targetable mutation in EGFR, indicating the limitation of target therapy for lung cancer. Moreover, 10.7–67.1% clonal mutations occurred after WGD illustrating that the WGD was a frequently truncal event in GGNs, consistent with findings from a non-small cell lung cancer study (41).
We identified two significantly mutated genes, EGFR and RBM10, across the GGNs, which were previously identified as potential drivers in LUAD (43). EGFR mutations were the most frequently observed in other studies of ground-glass nodular LUAD (60-62). RBM10 encodes an RNA-binding protein, and is subject to recurrent inactivating mutations in LUADs (43). A previous study in pancreatic ductal adenocarcinoma found that RBM10 mutations were associated with longer survival despite histological features of aggressive disease (63), subsequently, the high frequency of RBM10 mutations might be related to good prognosis of the patients with GGNs. Notably, the GGNs with RBM10 mutations tended to accompany a pathologically lepidic pattern (Fisher’s exact test, P<0.05), indicating better outcomes of patients with lepidic tumors than nonlepidic tumors. In agreement with our finding, patients with lepidic-predominate tumors showed better overall survival than patients with nonlepidic-predominate tumors (64-66). Our analysis of CNAs revealed significantly altered regions, including 5 known regions in LUAD, as well as two novel recurrent deletions at 3q29 and 9q13. Significantly amplified genes were overrepresented in functional terms associated with tumorigenesis, indicating the oncogenic potential of CNAs in the formation of GGNs. These putative drivers could serve as potential therapeutic targets to facilitate clinical therapy.
Mutations arising during the carcinogenesis of GGNs tended to accumulate in a clock-like manner, whereas the process of defective DNA mismatch repair was largely associated with genetic heterogeneity within GGNs. In agreement with recent studies in LUAD and melanoma sequencing studies (39,40,67), we also detected diverse mutational signatures during GGN progression. The clonal (trunk) mutations accumulated in a clock-like manner, whereas the reduction in clock-like signature 5 was observed in branch mutations. In the subclonal mutations, the signature associated with defective DNA mismatch repair was significantly dominant. None of the patients in this study received any systemic treatment prior to surgical removal of tumors; therefore, the switch in mutational processes was probably due to evolutionary changes occurring during GGN progression.
An unexpected observation was the high ITH within GGNs, as all the patients in this study were diagnosed with early-stage LUAD. In addition, relatively few putative driver mutations have been identified in individual tumors (Figure 1C). Although our research cannot show all the intermediate states before the observed ITH, it indeed indicated the evolutionary trajectory of GGNs. That is, on the background of low-mutation rates, a tumor-initiating cell population acquired mutations in a clock-like manner, and once a driver occurred, i.e., EGFR or RBM10 mutation, it was sufficient to allow a rapid expansion to produce a high ITH and numerous intermixed subclones, as suggested by the big bang model in colorectal cancer (68). Relatively short trunks (a mean of 25.8% clonal mutations) and early diversification observed in GGNs were more likely due to a single expansion rather than selective sweeps, which may reflect the early stage of carcinogenesis in LUAD. However, long-term progress of GGNs might further result in the acquisition of new driver mutations followed by selective sweeps and large clonal expansions. In this scenario, the tumor population could exhibit a decrease in ITH, as ~30% ITH was observed in other lung cancer studies (39-41).
The association between genetic variations and clinical characteristics were observed in this study. Mutation burden was highly associated with both solid portions and lepidic growth pattern (Figure 5A,B), and significantly differences in mutation burden was observed between mixed GGNs with a non-LEP pattern and pure GGNs with an LEP pattern (Figure 5C). We also observed a trend toward significantly higher clonal neoantigen burden in non-LEPs than in LEPs (Figure 5D). Both of mutation burden and clonal neoantigen burden could influence the response of patients to immune checkpoint inhibitors (55,56). Our results suggested nonlepidic-predominate lung cancer might be more sensitive to checkpoint blockade immunotherapy than lepidic-predominate lung cancer. Moreover, non-LEPs harbored significantly higher genomic instability than that of LEPs, which was characterized by GII scores (Figure 3C, P=0.041, Wilcoxon test), indicating a higher degree of malignancy in non-LEPs than in LEPs (45,46). Previous studies suggested that nonlepidic tumors were more malignant than lepidic tumors because patients with lepidic-predominate tumors showed better overall survival than patients with nonlepidic-predominate tumors (64-66), which was in agreement with the higher genomic instability observed in non-LEPs than in LEPs. Although the GGNs with RBM10 mutations tended to accompany a pathologically lepidic pattern (Fisher’s exact test, P=0.049), there is no evidence that RBM10 was associated with solid portions (Fisher’s exact test, P=1). Previous studies reported patients with lepidic-predominate tumors showed better overall survival than patients with nonlepidic-predominate tumors (64-66), which highlighted RBM10 may drive the distinct subtype of lung cancer with better prognosis. Moreover, we did not observed EGFR was associated with either solid portions (Fisher’s exact test, P=0.69) or lepidic pattern (Fisher’s exact test, P=1).
The main limitations of this study are (I) small sample size and (II) relative low tumor purity among GGN samples. The purpose of this study is trying to interrogate the subtle genetic changes in very early stage of lung cancer so we selected GGN samples with solid part of the nodules occupying less than 50% for special purpose, and spontaneously, pure GGN or mixed GGN are with low tumor purity inevitably. Only 28 patients who met our sample criterion were selected from our lung cancer cohort, and the small sample size decreased the statistic power during the stratified analysis so more samples are required for further validation against the results of this study.
In summary, we performed the first comprehensive genomic analysis for GGNs, providing insights into early events, frequent alterations and mutational processes during GGN evolution; we also determined genetic differences among clinical subtypes. Studies will move toward systematically integrating myriad of aspects of the GGN genome, including the interrelationships among multiple molecular levels, the interplay between somatic and germline variations as well as the tumor microenvironment and the immune system.
Data policy
The sequence data reported in this paper have been deposited in the Genome Sequence Archive (69) in BIG Data Center (70), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession numbers HRA000044 that are publicly accessible at http://bigd.big.ac.cn/gsa-human.
Acknowledgments
Funding: This study was supported by National Key R&D Program of China (Grant No. 2018YFC0910700), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB13000000), the Key Research Program of the Chinese Academy of Sciences (Grant No. KFZD-SW-220-1), the Natural Science Foundation of China (Grant Nos. 91531305, 31771416), the Innovation Program of the Beijing institute of genomics, Beijing Municipal Science & Technology Commission (Grant No. Z161100000516063), Special Fund of Beijing Municipal Administration of Hospitals Clinical Medicine Development (Grant No. XMLX201841), Beijing Human Resources and Social Security Bureau (Beijing Millions of Talents Project, Grant No. 2018A05), Peking University Medicine Seed Fund for Interdisciplinary Research (Grant No. BMU2018MX008), Beijing Natural Science Foundation (Grant No. 7192036), Beijing Municipal Administration of Hospitals’Youth Programme (Grant No. QML20171103).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-1086
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-1086
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-1086). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Human Research Ethics Committee of Beijing Cancer Hospital and Beijing Institute of Genomics (2015KT71), and informed consent was obtained from the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Allemani C, Matsuda T, Di Carlo V, et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 2018;391:1023-75. [Crossref] [PubMed]
- Webb WR. High resolution lung computed tomography. Normal anatomic and pathologic findings. Radiol Clin North Am 1991;29:1051-63. [PubMed]
- Muller NL. Differential diagnosis of chronic diffuse infiltrative lung disease on high-resolution computed tomography. Semin Roentgenol 1991;26:132-42. [Crossref] [PubMed]
- Oh JY, Kwon SY, Yoon HI, et al. Clinical significance of a solitary ground-glass opacity (GGO) lesion of the lung detected by chest CT. Lung Cancer 2007;55:67-73. [Crossref] [PubMed]
- Kim HY, Shim YM, Lee KS, et al. Persistent pulmonary nodular ground-glass opacity at thin-section CT: histopathologic comparisons. Radiology 2007;245:267-75. [Crossref] [PubMed]
- Henschke CI, Yankelevitz DF, Mirtcheva R, et al. CT screening for lung cancer: frequency and significance of part-solid and nonsolid nodules. AJR Am J Roentgenol 2002;178:1053-7. [Crossref] [PubMed]
- Chang B, Hwang JH, Choi YH, et al. Natural history of pure ground-glass opacity lung nodules detected by low-dose CT scan. Chest 2013;143:172-8. [Crossref] [PubMed]
- Detterbeck FC, Marom EM, Arenberg DA, et al. The IASLC Lung Cancer Staging Project: Background Data and Proposals for the Application of TNM Staging Rules to Lung Cancer Presenting as Multiple Nodules with Ground Glass or Lepidic Features or a Pneumonic Type of Involvement in the Forthcoming Eighth Edition of the TNM Classification. J Thorac Oncol 2016;11:666-80.
- Min JH, Lee HY, Lee KS, et al. Stepwise evolution from a focal pure pulmonary ground-glass opacity nodule into an invasive lung adenocarcinoma: an observation for more than 10 years. Lung Cancer 2010;69:123-6. [Crossref] [PubMed]
- Naidich DP, Bankier AA, MacMahon H, et al. Recommendations for the management of subsolid pulmonary nodules detected at CT: a statement from the Fleischner Society. Radiology 2013;266:304-17. [Crossref] [PubMed]
- Engeler CE, Tashjian JH, Trenkner SW, et al. Ground-glass opacity of the lung parenchyma: a guide to analysis with high-resolution CT. AJR Am J Roentgenol 1993;160:249-51. [Crossref] [PubMed]
- Qiu ZX, Cheng Y, Liu D, et al. Clinical, pathological, and radiological characteristics of solitary ground-glass opacity lung nodules on high-resolution computed tomography. Ther Clin Risk Manag 2016;12:1445-53. [Crossref] [PubMed]
- Shimada Y, Saji H, Otani K, et al. Survival of a surgical series of lung cancer patients with synchronous multiple ground-glass opacities, and the management of their residual lesions. Lung Cancer 2015;88:174-80. [Crossref] [PubMed]
- MacMahon H, Naidich DP, Goo JM, et al. Guidelines for Management of Incidental Pulmonary Nodules Detected on CT Images: From the Fleischner Society 2017. Radiology 2017;284:228-43. [Crossref] [PubMed]
- Moon Y, Sung SW, Lee KY, et al. Clinicopathological characteristics and prognosis of non-lepidic invasive adenocarcinoma presenting as ground glass opacity nodule. J Thorac Dis 2016;8:2562-70. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589-95. [Crossref] [PubMed]
- DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491-8. [Crossref] [PubMed]
- Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature Biotechnology 2013;31:213-9. [Crossref] [PubMed]
- Gerstung M, Papaemmanuil E, Campbell PJ. Subclonal variant calling with multiple samples and prior knowledge. Bioinformatics 2014;30:1198-204. [Crossref] [PubMed]
- Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568-76. [Crossref] [PubMed]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164 [Crossref] [PubMed]
- Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011;27:2987-93. [Crossref] [PubMed]
- Favero F, Joshi T, Marquard AM, et al. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann Oncol 2015;26:64-70. [Crossref] [PubMed]
- Nahar R, Zhai W, Zhang T, et al. Elucidating the genomic architecture of Asian EGFR-mutant lung adenocarcinoma through multi-region exome sequencing. Nat Commun 2018;9:216. [Crossref] [PubMed]
- Dewhurst SM, McGranahan N, Burrell RA, et al. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discov 2014;4:175-85. [Crossref] [PubMed]
- McGranahan N, Favero F, de Bruin EC, et al. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci Transl Med 2015;7:283ra54 [Crossref] [PubMed]
- Blokzijl F, Janssen R, van Boxtel R, et al. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med 2018;10:33. [Crossref] [PubMed]
- Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214-8. [Crossref] [PubMed]
- Martincorena I, Raine KM, Gerstung M, et al. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell 2018;173:1823. [Crossref] [PubMed]
- Mermel CH, Schumacher SE, Hill B, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011;12:R41. [Crossref] [PubMed]
- Dennis G Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003;4:3. [Crossref] [PubMed]
- Shukla SA, Rooney MS, Rajasagi M, et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat Biotechnol 2015;33:1152-8. [Crossref] [PubMed]
- Jurtz V, Paul S, Andreatta M, et al. NetMHCpan-4.0: Improved Peptide-MHC Class I Interaction Predictions Integrating Eluted Ligand and Peptide Binding Affinity Data. J Immunol 2017;199:3360-8. [Crossref] [PubMed]
- Yang Y, Yang Y, Zhou X, et al. EGFR L858R mutation is associated with lung adenocarcinoma patients with dominant ground-glass opacity. Lung Cancer 2015;87:272-7. [Crossref] [PubMed]
- Lederlin M, Puderbach M, Muley T, et al. Correlation of radio- and histomorphological pattern of pulmonary adenocarcinoma. Eur Respir J 2013;41:943-51. [Crossref] [PubMed]
- . Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Luo W, Tian P, Wang Y, et al. Characteristics of genomic alterations of lung adenocarcinoma in young never-smokers. Int J Cancer 2018;143:1696-705. [Crossref] [PubMed]
- de Bruin EC, McGranahan N, Mitter R, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 2014;346:251-6. [Crossref] [PubMed]
- Zhang J, Fujimoto J, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 2014;346:256-9. [Crossref] [PubMed]
- Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2109-21. [Crossref] [PubMed]
- Sordella R, Bell DW, Haber DA, et al. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science 2004;305:1163-7. [Crossref] [PubMed]
- Devarakonda S, Morgensztern D, Govindan R. Genomic alterations in lung adenocarcinoma. Lancet Oncol 2015;16:e342-51. [Crossref] [PubMed]
- Cairo S, Armengol C, De Reynies A, et al. Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell 2008;14:471-84. [Crossref] [PubMed]
- Schneider BL, Kulesz-Martin M. Destructive cycles: the role of genomic instability and adaptation in carcinogenesis. Carcinogenesis 2004;25:2033-44. [Crossref] [PubMed]
- Duesberg P, Li R. Multistep carcinogenesis: a chain reaction of aneuploidizations. Cell Cycle 2003;2:202-10. [Crossref] [PubMed]
- Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature 2007;450:893-8. [Crossref] [PubMed]
- Zack TI, Schumacher SE, Carter SL, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet 2013;45:1134-40. [Crossref] [PubMed]
- Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010;463:899-905. [Crossref] [PubMed]
- Nymark P, Wikman H, Ruosaari S, et al. Identification of specific gene copy number changes in asbestos-related lung cancer. Cancer Res 2006;66:5737-43. [Crossref] [PubMed]
- Yoon KA, Park JH, Han J, et al. A genome-wide association study reveals susceptibility variants for non-small cell lung cancer in the Korean population. Hum Mol Genet 2010;19:4948-54. [Crossref] [PubMed]
- Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 2012;30:413-21. [Crossref] [PubMed]
- Fujiwara T, Bandi M, Nitta M, et al. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 2005;437:1043-7. [Crossref] [PubMed]
- Kakinuma R, Ohmatsu H, Kaneko M, et al. Progression of focal pure ground-glass opacity detected by low-dose helical computed tomography screening for lung cancer. J Comput Assist Tomogr 2004;28:17-23. [Crossref] [PubMed]
- McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016;351:1463-9. [Crossref] [PubMed]
- Hellmann MD, Nathanson T, Rizvi H, et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell 2018;33:843-52.e4. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Chabanon RM, Pedrero M, Lefebvre C, et al. Mutational Landscape and Sensitivity to Immune Checkpoint Blockers. Clin Cancer Res 2016;22:4309-21. [Crossref] [PubMed]
- Li R, Li X, Xue R, et al. Early metastasis detected in patients with multifocal pulmonary ground-glass opacities (GGOs). Thorax 2018;73:290-2. [Crossref] [PubMed]
- Kobayashi Y, Mitsudomi T, Sakao Y, et al. Genetic features of pulmonary adenocarcinoma presenting with ground-glass nodules: the differences between nodules with and without growth. Annals of Oncology 2015;26:156-61. [Crossref] [PubMed]
- Lee H, Joung JG, Shin HT, et al. Genomic alterations of ground-glass nodular lung adenocarcinoma. Sci Rep 2018;8:7691. [Crossref] [PubMed]
- Park E, Ahn S, Kim H, et al. Targeted Sequencing Analysis of Pulmonary Adenocarcinoma with Multiple Synchronous Ground-Glass/Lepidic Nodules. J Thorac Oncol 2018;13:1776-83. [Crossref] [PubMed]
- Witkiewicz AK, McMillan EA, Balaji U, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun 2015;6:6744. [Crossref] [PubMed]
- Warth A, Muley T, Meister M, et al. The novel histologic International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification system of lung adenocarcinoma is a stage-independent predictor of survival. J Clin Oncol 2012;30:1438-46. [Crossref] [PubMed]
- Yoshizawa A, Sumiyoshi S, Sonobe M, et al. Validation of the IASLC/ATS/ERS lung adenocarcinoma classification for prognosis and association with EGFR and KRAS gene mutations: analysis of 440 Japanese patients. J Thorac Oncol 2013;8:52-61. [Crossref] [PubMed]
- Tsao MS, Marguet S, Le Teuff G, et al. Subtype Classification of Lung Adenocarcinoma Predicts Benefit From Adjuvant Chemotherapy in Patients Undergoing Complete Resection. J Clin Oncol 2015;33:3439-46. [Crossref] [PubMed]
- Harbst K, Lauss M, Cirenajwis H, et al. Multiregion Whole-Exome Sequencing Uncovers the Genetic Evolution and Mutational Heterogeneity of Early-Stage Metastatic Melanoma. Cancer Res 2016;76:4765-74. [Crossref] [PubMed]
- Sottoriva A, Kang H, Ma Z, et al. A Big Bang model of human colorectal tumor growth. Nat Genet 2015;47:209-16. [Crossref] [PubMed]
- Wang Y, Song F, Zhu J, et al. GSA: Genome Sequence Archive Genomics Proteomics Bioinformatics 2017;15:14-8. [Crossref] [PubMed]
- BIG Data Center Members. The BIG Data Center: from deposition to integration to translation. Nucleic Acids Res 2017;45:D18-D24. [Crossref] [PubMed]


