
This article has an erratum available at: http://dx.doi.org/10.21037/tlcr-2021-4 the article has been update on 2021-09-27 at here.
Tumor-associated macrophages secret exosomal miR-155 and miR-196a-5p to promote metastasis of non-small-cell lung cancer
Introduction
According to global cancer statistics in 2018, lung cancer accounts for ~12% of newly diagnosed cancer cases and ~18% of total cancer deaths (1). About 85% of diagnosed lung cancer is non-small cell lung cancer (NSCLC), which have a low 5-year survival rate (<15%) (2,3). Although timely treatments such as surgery, chemotherapy and radiotherapy can prolong the overall survival of patients, NSCLC is still the most fatal disease. The causes of lung cancer death vary and metastasis is a predominant factor (4). Therefore, understanding the molecular basis of NSCLC metastasis is critical for proposing new therapeutic approaches for its treatment (5-7). Several classes of biomolecules, such as microRNAs (miRNAs), matrix metalloproteinases and transcription regulatory protein BACH1, have been well studied in the metastasis of NSCLC (8-11). Although tumor microenvironment is also known to mediate NSCLC metastasis, the role of key components of tumor microenvironment in NSCLC metastasis has not been fully elucidated (12,13).
The tumor microenvironment consists of cancer cells and stromal cells that include macrophages, endothelial cells and fibroblasts (14-16). Moreover, products of these cells, such as cytokines, growth factors and enzymes, are also present in the tumor microenvironment. Among these components in the tumor microenvironment, tumor-associated macrophages (TAMs) are the pivotal orchestrators as a class of immune cells (17). TAMs have two populations, M1 and M2 types. It has been found that M1 TAMs have an anti-tumor function, whereas M2 TAMs can promote tumor metastasis (18). For NSCLC, M1 TAMs can suppress the angiogenesis and are positively related with the survival time of NSCLC patients (19,20). In contrast, M2 TAMs are found to promote NSCLC metastasis through activating the epithelial-mesenchymal transition (EMT) of NSCLC cells and the invasion of lung cancer cells (20,21). However, the detailed mechanisms by which M2 TAMs promote NSCLC metastasis remain elusive.
Exosomes are small vesicles secreted by parental cells into extracellular microenvironment to mediate cross-talk between parental cells and target cells (22,23). Almost all cell types have the capacity to release exosomes and the size of exosomes is ~30–150 nm in diameter. To mediate cell-cell communication, exosomes deliver proteins, miRNAs or mRNAs into target cells to change the gene expression (24). For instance, gastric cancer cells secret exosomes carrying miR-21 into peritoneal mesothelial cells to promote peritoneal metastasis (22). For the cross-talk between cancer cells and TAMs, exosomes carrying miRNAs are also key players. Taking an example, ovarian cancer cells secret exosomes containing miR-222 to induce M2 polarization of TAMs (25). Moreover, M2 TAMs also can release exosomes carrying miR-501 to promote the progression of pancreatic cancer (26). Nevertheless, whether M2 TAMs promote NSCLC metastasis through exosomes and miRNAs remains unclear.
Ras association domain family member 4 (RASSF4), a member of the RASSF family, abnormally expresses in human cancers and involves in the carcinogenesis process. Moreover, it had a suppression effect on the biological process. For instance, in osteosarcoma cells, RASSF4 overexpression markedly inhibits proliferation, invasion and EMT (27). In NSCLCs, it is reported that RASSF4 is down-regulated in NSCLC tissues and effectively inhibits cancer cell proliferation and invasion (28). These studies suggested that RASSF4 serves as an important tumor suppressor in the development of cancer.
Here, we report that M2 TAMs promote EMT of NSCLC cells through secreting exosomes. MiR-155 and miR-196a-5p are the key functional molecule in M2 TAMs-released exosomes that promote EMT of NSCLC cells through targeting RASSF4.
We present the following study in accordance with the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-1255.
Methods
Patients
NSCLC localized tissues (n=15) and metastatic tissues (n=15) were obtained from patients in Jiangsu Province People’s Hospital between 2015 and 2016. The project has attained informed consent from all patients and was approved by the ethics committee of Jiangsu Province People’s Hospital and the First Affiliated Hospital of Nanjing Medical University (No. 2019-SR-266). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell lines and cell culture
The A549 and THP-1 cells were obtained from Chinese Academy of Sciences, Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). A549/Luc cells (A549 cells stably express luciferase) were constructed by Synthgene (Nanjing, China). THP-1 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium and A549 cells were cultured in DMEM (Dulbecco’s Modified Eagle’s Medium) (Gibco, Rockville, USA) supplemented with 10% FBS (fetal bovine serum) (Gibco, Rockville, USA) and 100 µg/mL streptomycin and penicillin (Gibco, Rockville, USA) in a humidified atmosphere at 37 °C. After 7 days in culture, THP-1 cells were treated with 100 ng/mL phorbol 12-myristate-13-acetate (PMA) to differentiate into macrophages for 48 h. Following treatment with 20 ng/mL interleukin-4 (IL-4) for 72 h, the cells were polarized into M2 macrophages.
Cell transfection
MiR-155 inhibitor, miR-155 mimic, miR-196a-5p inhibitor, miR-196a-5p mimic, short hairpin (sh-RASSF4) and negative control (NC) were synthesized in Gene Pharma Company (Shanghai, China). Lipofectamine 2000 (Invitrogen, USA) was performed for cell transfection based on the specification.
Immunohistochemistry assay
The immunohistochemistry assay was performed according to Yin et al. (26). The sections were incubated with the primary antibody of CD68 (Ab955, 1: 100, Abcam, Cambridge, UK) at 4 °C overnight and horseradish peroxidase labeled goat anti-mouse IgG antibody (A205719, 1:200, Abcam, Cambridge, UK) at room temperature for 1 h. The color reaction was performed with diaminobenzidine chromogen solution (Dako, Carpinteria, USA). Brown-yellow particles represented the positive expression of CD68 protein and the blue particles represented the nucleus stained by hematoxylin (Sigma, USA).
Flow cytometry assay
The macrophages were extracted by the tumor macrophage isolation fluid (Sangon Biotch, China). Then, macrophages were stained with CD163 (Abcam, USA) and CD206 (Abcam, USA) for 30 min at 4 °C. The labeled cells were analyzed by FACScan flow cytometry (BD Biosciences, USA).
RNA extraction and quantitative real-time PCR analysis
The extraction and reverse transcription of total RNA were performed according to the previous report (29). The expression levels of TNF-α, IRF5, IRF4, Arg-1 and miR-155 were analyzed by quantitative real-time PCR with the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene or U6 as a standard control. Primers of TNF-α, IRF5, IRF4, Arg-1, miR-155, miR-196a-5p, miR-3091-3p, miR-12206-5p, miR-12180-3p, U6 and GAPDH were as follows: TNF-α (Forward: 5'-CCTCTCTCTAATCAGCCCTCTG-3'; Reverse: 5'-GAGGACCTGGGAGTAGATGAG-3'); IRF5 (Forward: 5'-GGGCTTCAATGGGTCAACG-3'; Reverse: 5'-GCCTTCGGTGTATTTCCCTG-3'); IRF4 (Forward: 5'-GCTGATCGACCAGATCGACAG-3'; Reverse: 5'-CGGTTGTAGTCCTGCTTGC-3'); Arg-1 (Forward: 5'-GTGGAAACTTGCATGGACAAC-3'; Reverse: 5'-AATCCTGGCACATCGGGAATC-3'); miR-155 (Forward: 5'-GGAGGTTAATGCTAATCGTGATAG-3; Reverse: 5'-GTGCAGGGTCCGAGGT-3'); miR-196a-5p (Forward: 5'-CCGACGTAGGTAGTTTCATGTT-3; Reverse: 5'-GTGCAGGGTCCGAGGTATTC-3'); miR-3091-3p (Forward: 5'-GCGGGCCTGACCAGTCT-3; Reverse: 5'-AGTGCAGGGTCCGAGGTATT-3'); miR-12206-5p (Forward: 5'-GCGCGTACTATGCCTGGAAG-3; Reverse: 5'-AGTGCAGGGTCCGAGGTATT-3'); miR-12180-3p (Forward: 5'-GCGCGAGGAGCTGTGGA-3; Reverse: 5'-AGTGCAGGGTCCGAGGTATT-3'); U6 (Forward: 5'-TCGGCAGCACATATACTAA-3'; Reverse: 5'-CGCTTCACGAATTTGCGTGT-3'); GAPDH (Forward: 5'-GACCTCAACTACATGGTT-3'; Reverse: 5'-AACCATGTAGTTGAGG-3'). These primers were synthesized and purified by RiboBio (Guangzhou, China).
CCK-8 assay
The cell viability was measured by CCK-8 assay (Beyotime, China). A549 cells (1×104 cells/well) were cultured in 96-well plates and cultured for 24, 48 and 72 h. Subsequently, 10 µL of CCK-8 reagent was incubated for additional 4 h at 37 °C. At last, the cell optical density was detected by a Microplate Reader (Bio-Rad, USA) with absorbance at 450 nm.
Cell migration and invasion assays
The cell invasion and migration assays were performed by 24-well Transwell cell culture chambers with 8-µm sized pores with or without precoated Matrigel (BD Biosciences, San Jose, CA, USA). Specifically, A549 cells, A549 cells co-cultured with M2 macrophages treated with or without GW4869 and A549 cells co-cultured with exosomes from M0/M2 macrophages or M2 macrophages transfected with different plasmids, at a density of 5×104 cells/mL, were re-suspended with 200 µL DMEM medium (serum-free) and seeded into the upper chamber, while the lower chamber was placed with 600 µL DMEM medium (10% FBS). After incubation for 24 h, the cells remaining in the upper chamber were removed, the invaded or migrated A549 cells were fixed with the methanol (100%), stained with crystal violet (0.1 mg/mL) and counted under a microscope.
Isolation, identification and labeling of exosomes
The exosomes from M0 or M2 macrophages were isolated by density gradient ultracentrifugation according to previously reported protocol (30). Briefly, cell culture medium was collected and centrifuged at 1,000 g for 10 min, 2,000 g for 20 min, 4,000 g for 30 min and 10,000 g for 1 h to obtain the supernatant. The exosomes were collected by centrifuging the supernatant at 100,000 g for 2 h at 4 °C. The size distribution and concentration of exosomes were analyzed at a flow rate of 0.03 Ml per min using a Zetasizer Nano ZS (Malvern Instrument, UK) and NanoSight NS300 (Westborough, MA, USA), respectively. Purified exosomes were labeled with the PKH-67 green fluorescent linker mini kit (Sigma, USA) according to the manufacturer’s instructions.
miRNA profiling
The aberrant miRNAs expressions of M0-exosome and M2-exosome were analyzed by miRNAs sequencing. Briefly, total RNA was isolated from M0-exosome and M2-exosome using TRIzol reagent (Invitrogen, MA, USA). 250 ng total RNA of M0-exosome and M2-exosome were extracted to prepare the small RNA sequencing library by using the NEBNext Multiplex Small RNA Library Prep Set for Illumina (NEB, USA). The libraries were finally sequenced and the Solexa CHASTITY quantity filtered reads were harvested as Clean Reads. For data analysis, differentially expressed miRNA profiles between M0-exosome and M2-exosome groups were compared, fold changes were calculated to identify significant differentially expressed miRNAs and hierarchical clustering was performed. The selected miRNAs were verified by qRT-PCR.
Western blot analysis
The radio-immunoprecipitation assay (RIPA) lysis buffer (Solarbio, Shanghai, China) with 0.5% phenylmethanesulfonyl fluoride (PMSF) (Solarbio, Shanghai, China) was used to extract the total protein of exosomes, cells or tissues. The protein concentration was quantified by the BCA protein quantification Kit (Sigma, USA). The primary antibodies in this study were purchased from Abcam: rabbit anti-CD63 (ab68418, 1:500), CD81 (ab109201, 1:200), RASSF4 (ab243709, 1:1,000) and TSG101 (ab30871, 1:500); mouse anti-E-cadherin (ab11512, 1:1,000), vimentin (ab8978, 1:1,000) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (ab8245, 1:5,000). The horseradish peroxidase labeled goat anti-rabbit IgG antibody (ab205718, 1:10,000) and goat anti-mouse antibody (ab6789, 1:10,000) were available as the secondary antibodies. Image J software was used to quantify each protein band.
Luciferase reporter assay
The 3'-UTR segments of RASSF4 in wild type and mutant were synthesized and inserted into a firefly luciferase reporter construct. Cells were co-transfected with RASSF4 construct and miR-155 mimic or miR-196a-5p mimic. Luciferase activity in this study was measured by the Dual-Luciferase Reporter Assay System (Promega, USA) according to the protocol.
Animal studies
Six-week-old male athymic BALB/c nude mice (15–20 g) were purchased from the Model Animal Research Center of Nanjing University (Nanjing, China) and housed in a temperature-controlled room with a 12-hour light/dark cycle. Diet and water were available ad libitum. For the in vivo lung metastases model, exosomes purified from M2 macrophages or M2 macrophages transfected with 1×109 ifu of miR-155 inhibitor lentivirus or M2 macrophages transfected with 1×109 ifu of miR-196a-5p inhibitor lentivirus were respectively injected into the peritoneum. Four days post-injection, A549/Luc cells were injected into the tail vein of representative mice (n=5 per group, total 25). All mice were grouped randomly. The luciferase signal intensity from days 0 to 28 is on equivalent scales in the models. Bioluminescent flux (photons/s/cm2/steradian) was determined for the lung metastases. Metastatic progression was monitored and imaged using an IVIS-100 system (Caliper Life Sciences, MA, USA) 10 min after intraperitoneal injection of luciferin (300 mg/kg i.v.) in 80 µL of saline. After 28 days, mice were euthanized using large doses of intraperitoneal pentobarbital (200 mg/kg) in the laboratory and tissues were separated for further experiments. Animal care and euthanasia were carried out with the approval of the Institutional Animal Care and Use Committee (IACUC) of Nanjing Medical University, in compliance with the national or institutional guidelines for the care and use of animals.
Hematoxylin & eosin (HE) staining
The dewaxed sections were firstly incubated with hematoxylin to stain the nucleus for 5 min, then 1% ethanol-hydrochloric acid for 30 s and eosin solution for 3 min. Finally, the sections were dehydrated in graded alcohol following by clearing in xylene.
Statistical analysis
All statistical analyses were performed by GraphPad Prism 6.0 software. The Student’s t-test was used to analyze significant differences in this study. The error bars indicate the standard deviation from the mean of triplicate measurements. Asterisks indicate significant differences (*P<0.05; **P<0.01; ***P<0.001) compared with the corresponding control.
Results
M2 TAMs are abundant in metastatic tissues of NSCLC patients
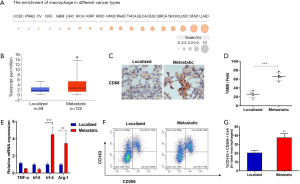
We firstly analyzed the enrichment of macrophage in different cancer types through the online database TCIA (https://tcia.at/). The result suggested that macrophages were most abundant in lung cancer (Figure 1A). NSCLC accounts for more than 80% of lung cancer and metastasis causes 70% of deaths. Then, to evaluate the role of macrophages in the metastasis of NSCLC, we utilized gene set enrichment analysis via the Cancer Genome Atlas database and observed that CD68, a macrophage marker, was significantly increased in 122 patients with the metastasis of NSCLC in Figure 1B. To further validate the distribution of macrophages in NSCLC, we analyzed the expression of CD68 by immunohistochemical staining and found that the higher density of TAMs in NSCLC metastasis tissues than that in localized tissues (Figure 1C,D), suggesting TAMs recruitment related to the metastasis of NSCLC. Next, we estimated the two subpopulations of macrophages including macrophage M0 and M2 in NSCLC patients. The results of qRT-PCR showed that the transcription expressions of M1 macrophages marker such as tumor necrosis factor alpha (TNF-α) and interferon regulatory factor 5 (IRF5) had a significant decrease in all localized samples in this study; however, the signatures of M2 macrophages like interferon regulatory factor 4 (IRF4) and arginase-1 (Arg-1) were up-regulated in metastatic tissues compared with localized tissues (Figure 1E). Moreover, flow cytometry assay suggested that M2 macrophages were more abundant in metastatic tissues (Figure 1F,G). These data indicated that M2 TAMs are probably involved in the metastasis of NSCLC.
M2 TAMs promote migration, invasion and EMT of A549 cells
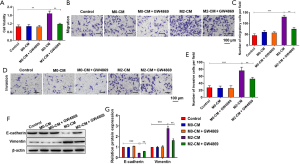
The results in Figure 1 showed that M2 macrophages were more enriched in metastatic NSCLC patients than those without metastasis. To evaluate whether TAMs were involved in the metastasis of NSCLC, we co-cultured A549 cells with M0 or M2 macrophages in a Transwell insert. The macrophages have been extensively reported to regulate a series of biological processes of NSCLC through exosomes. To evaluate whether the effect of macrophages on A549 cells was through exosomes, we treated M0 or M2 macrophages with GW4869 (an inhibitor of exosome release). For inhibition of exosome generation, macrophages were cultured in media containing 10 µM GW4869. CCK-8 assay displayed that M2 macrophages significantly promoted cell viability compared to the control, and GW4869 treatment reversed the accumulation of cell viability caused by M2 macrophages (Figure 2A). The results of Transwell assay unveiled that the migration and invasion capacity of A549 cells was enhanced in A549 cells co-cultured with M2 macrophages group compared with the control, suggesting that M2 macrophages can promote the migration and invasion potential of A549 cells (Figure 2B,C,D,E). Following GW4869 treatment, M2 macrophages failed to promote the migration and invasiveness of A549 cells, suggesting that exosomes derived from M2 macrophages play an essential role in the migration and invasiveness of A549 cells (Figure 2B,C,D,E). Besides, M2 macrophages decreased the protein expression of epithelial cell marker (E-cadherin) and promoted the expression of mesenchymal cell marker (vimentin), and GW4869 treatment reversed the effects (Figure 2F,G). Above all, these results clarified that M2 macrophages markedly promoted migration, invasion, and EMT of A549 cells.
Exosomes secreted by M2 TAMs promote migration, invasion and EMT of A549 cells
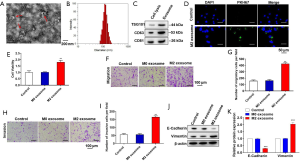
Subsequently, a large number of exosomes were found in the supernatant of M2 macrophages culture by transmission electron microscope. And the shape and size of exosomes were solid with typical structure of two-layer membrane and exosomes had an average diameter of 100 nm as shown in Figure 3A,B. Western blot analysis in Figure 3C showed that the protein levels of exosomal markers TSG101, CD63 and CD81 had a significant increase in exosomes compared with cell lysis, confirming the successful extraction of exosomes. To examine whether M2 macrophages derived exosomes (M2 exosomes) can be taken up by A549 cells, we pre-labeled M2 exosomes with PKH67. In Figure 3D, A549 cells co-cultured with PKH67-labeled M2 exosomes for 48 h exhibited green fluorescence, confirming the exosomes derived from M2 macrophages can be taken up by A549 cells. Furthermore, after 48 h of co-culture of A549 cells with M0 exosomes or M2 exosomes in vitro. CCK-8 assay, Transwell assay and western blot analysis were applied to examine whether exosomes generated from M2 macrophages were sufficient to induce the cell migration and invasion. CCK-8 assay suggested that exosomes secreted from M2 macrophages notably raised the A549 cell viability, while exosomes from M0 macrophages had no effect on cell viability (Figure 3E). The results in Figure 3F,G,H,I revealed that the invasiveness and migration ability of A549 cells co-cultured with exosomes secreted from M2 macrophages were strengthen compared with that co-cultured with M0 exosomes. Moreover, the protein expression levels of E-cadherin decreased and the expression levels of vimentin increased in A549 cells co-cultured with M2 exosomes in Figure 3J,K. These data clarified that exosomes secreted from M2 macrophages can markedly promote migration, invasion, and EMT of A549 cells.
M2 macrophages-derived exosomes promote migration, invasion and EMT of A549 cells through transporting miR-155 and miR-196a-5p
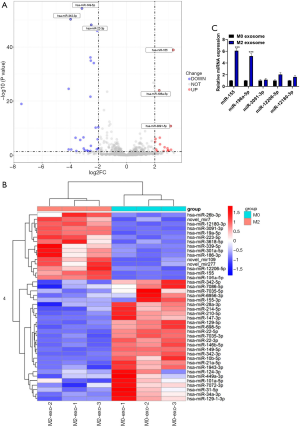
Emerging evidence indicated that miRNAs involved in cell-cell communications are frequently encapsulated in exosomes, and implement their biological functions in the recipient cells. To explore the mechanisms by which M2-exosome produced a marked effect in NSCLC, we generated miRNA profiles of M0-exosome and M2-exosome by miRNA microarray analysis. The comparison of the expression levels of the miRNAs in the M0-exosome and M2-exosome groups is depicted in Figure 4A,B. Among the miRNAs, miR-155 and miR-196a-5p were the most abundant in M2-exosome group in Figure 4A,B. In further analysis, in Figure 4C, miR-155 and miR-196a-5p were markedly upregulated in M2-exosome group. The results showed that miR-155 and miR-196a-5p were enriched in M2 macrophages-derived exosomes. Exosomes deliver proteins, miRNAs or mRNAs into target cells to mediate a series of physiological processes. Therefore, we hypothesized that M2-exosomes might induce the increased migration of recipient A549 cells through the transfer of functional miR-155 and miR-196a-5p. According to CCK-8 assay, we found that M2-exosomes promoted the cell viability, then the viability was restrained by transfecting miR-155 inhibitor and miR-196a-5p inhibitor (Figure 5A). In Figure 5B,C,D,E, we found that the M2-exosomes promoted the migration of A549 cells, increased the protein expression level of vimentin and inhibited the E-cadherin expression, while the opposite pattern could be observed in M2/miR-155-inhibitor exosomes group and M2/miR-196a-5p-inhibitor exosomes group. Taken together, these data suggested that miR-155 and miR-196a-5p carried by exosomes derived from M2 macrophages mediated a series of biological processes of NSCLC.
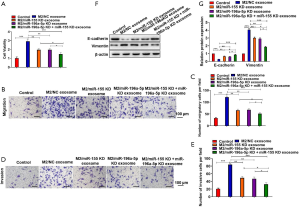
The direct target of exosomal miR-155 and miR-196a-5p in NSCLC is RASSF4
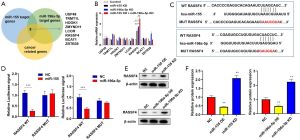
To explore the common targets between miR-155 and miR-196a-5p participated in the development of NSCLC, online bioinformatics database (TargetScan Release 7.0) was executed to predict the target genes. Based on these, David’s bioinformatics resources and literature review were performed to evaluate analyze genes function in cancer, which showed 8 genes were associated with cancer progression (USP48, TRMT1L, HOOK1, ZMYND11, LCOR, RASSF4, BCAT1, ZBTB39) (Figure 6A). Moreover, RT-qPCR analysis displayed that RASSF4 had the largest fold difference in A549 cells treating with M2 exosomes (Figure 6B). The prediction of TargetScan Release 7.0 database showed that RASSF4 in 3'-untranslated regions (UTRs) possesses putative binding sites for miR-155 and miR-196a-5p (Figure 6C). Subsequently, the luciferase reporter plasmids containing the wild type (WT) or mutation-type of 3'-UTR of RASSF4 were successfully transfected into A549 cells. As shown in Figure 6D, miR-155 or miR-196a-5p significantly decreased the luciferase activity of WT RASSF4 compared with NC. Western blot analysis revealed that miR-155 or miR-196a-5p overexpression significantly down-regulated the expression of RASSF4, while miR-155 or miR-196a-5p inhibitor up-regulated RASSF4 expression (Figure 6E,F). Collectively, these results testified that RASSF4 was the target of miR-155 and miR-196a-5p.
MiR-155 and miR-196a-5p promote migration, invasion and EMT of A549 cells through regulating RASSF4 expression
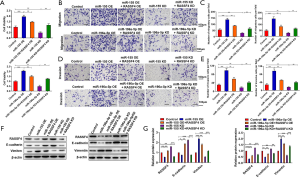
We carried out the rescue assays to further explore whether miR-155 and miR-196a-5p executed its performance by RASSF4. The result of CCK-8 assay indicated that overexpression of RASSF4 suppressed the cell viability induced by miR-155 mimic and miR-196a-5p mimic, while the repression of RASSF4 abrogated the anti-proliferation effects of miR-155 inhibitor and miR-196a-5p inhibitor (Figure 7A). Similar results were observed in cell migration and invasion (Figure 7B,C,D,E). Western blot analysis revealed that the significant down-regulation in the expression of RASSF4 and E-cadherin, and up-expression of vimentin resulting from miR-155 and miR-196a-5p overexpression, these results were reversed when RASSF4 was overexpressed. Correspondingly, the promoted expressions of RASSF4 and E-cadherin by miR-155 and miR-196a-5p knockdown were alleviated by RASSF4 inhibition (Figure 7F,G). Taken together, the effects of miR-155 and miR-196a-5p on A549 cells, including cell viability, cell migration, cell invasion, and the protein levels of RASSF4, vimentin and E-cadherin, could be reversed by RASSF4.
M2 TAMs secret exosomal miR-155 and miR-196a-5p to promote NSCLC metastasis in vivo
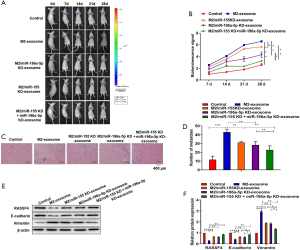
6-week-old healthy male BALB/c nude mice were performed to establish transplantation tumor model. To assess the effect of miR-155 and miR-196a-5p carried by exosomes from M2 macrophages on lung metastasis in vivo, M2 exosome, M2/miR-155 knockdown exosome, M2/miR-196a-5p knockdown exosome, M2/miR-155 knockdown + miR-196a-5p knockdown exosome were injected respectively after intravenous injection of A549/Luc cells into mice to construct lung metastasis model. In Figure 8A,B, M2-derived exosomes markedly contributed to the lung dissemination. And the high expression of luciferase activity in M2 exosome group could be effectively decreased through the knockdown of miR-155 and miR-196a-5p. Next, HE staining from lung tissues of each group in Figure 8C,D showed that M2-derived exosomes with the knockdown of miR-155 and miR-196a-5p markedly contributed to alleviate the metastasis of NSCLC resulting from M2 exosomes. In Figure 8E,F, western blot analysis from tumor tissues of each group revealed that the protein levels of RASSF4 in M2/miR-155 knockdown and miR-196a-5p knockdown exosome group were significantly up-regulated compared with M2-exosome group. Additionally, M2/miR-155 knockdown and miR-196a-5p knockdown exosome group promoted E-cadherin expression and decreased vimentin expression compared with M2 exosome group. In general, our findings reveal that NSCLC metastasis can be promoted by exosomal miR-155 and miR-196a-5p secreted from M2 TAMs in vivo.
Discussion
Due to the metastasis to bone, brain and liver, poor prognosis is common in lung cancer patients, resulting in high mortality (31). Although various biomolecules have been found to mediate lung cancer metastasis, knowledge regarding to roles of tumor microenvironment in lung cancer metastasis has largely lagged behind. Stromal cells including TAMs in the tumor microenvironment are either induced or directly interacting with cancer cells to promote the progression and metastasis of lung cancer (32,33). The modes of action of stromal cells include the secretion of extracellular components, such as growth factors, chemokines and exosomes (34). However, due to the extreme complexity of these processes, the molecular mechanisms underlying lung metastasis in the tumor microenvironment have not been fully understood.
Multiple evidence has expounded that M2 TAMs participated in the progression of tumors (35). In the study, we found that macrophages were most abundant in lung cancer, and M2 macrophages were rich in metastatic tissues. Based on these results, we speculated M2 TAMs were involved in the metastasis of NSCLC. The alveolar M2 TAM density was reported to be associated with tumor differentiation, invasive size, pathological stage and poor prognosis (36). The results indicated that M2 TAMs promoted NSCLC metastasis, which were consistent with Yuan and Guo (20,21). Nevertheless, the exact mechanisms by which M2 TAMs promote NSCLC metastasis remained undefined. There is increasing evidence that stromal exosomes play an important role in the tumor microenvironment as a medium for intercellular communication (37). For instance, Zheng et al. suggested that M2 TAMs-derived exosomes promoted migration of gastric cancer cells (38). Similarly, our result indicated that exosomes secreted from M2 macrophages can markedly promote migration, invasion, and EMT.
Exosomes from macrophages easily selectively deliver biologically active substances, such as proteins, miRNAs or mRNAs, to recipient cells to achieve efficient interflow between macrophages and tumor cells (39). MiRNAs are important gene regulators, which control various physiological and pathological processes (40). For instance, Yin et al. reported that exosomal miR-501-3p from macrophages induced the progression of pancreatic ductal adenocarcinoma through TGFBR3 (26). Besides, Lan et al. clarified that M2 macrophage-derived exosomes transferred miR-155-5p and miR-21-5p to colorectal cancer cells to accelerate tumor progression (41). In this paper, we demonstrated that the levels of miR-155 and miR-196a-5p were up-regulated in M2 TAMs-derived exosomes. MiR-155 is previously identified as an oncogenic miRNA in various cancers, such as breast cancer, ovarian cancer and lung cancer (42-44). Previous studies have also suggested that miR-19a-5p facilitated the cell proliferation in colorectal cancer (45), bladder cancer (46), gastric cancer (47). However, whether miR-196a-5p regulates the progression of NSCLC is not fully understood. In this paper, a series of gain-of-function and loss-of-function experiments illustrated exosomal miR-155 and miR-196a-5p promoted EMT and migration of NSCLC cells and promoted NSCLC metastasis in vivo.
It is confirmed that miRNAs regulate the biological effects of cancer cell by directly binding to the 3'UTR of mRNA (48). Therefore, targeting certain miRNAs involved in NSCLC has been considered as an efficient therapeutic approach (49). Moreover, we found that RASSF4 was a direct downstream target of both miR-155 and miR-196a-5p. RASSF4 is considered as a protein of the RASSF family (27). RASSF proteins directly interact with RAS proteins with effector characteristics to induce RAS-dependent cell death (50). In addition, RASSF protein has been reported to play an important role in tumor inhibition and is involved in many important biological functions, such as proliferation, cell cycle, apoptosis, and DNA repair (51). A number of studies suggested that RASSF4 was widely expressed in normal tissues but down-regulated in tumors, indicating that RASSF4 acted as a tumor suppressor in numerous malignancies (27,52). In our study, we found that exosomal miR-155 and miR-196a-5p secreted by M2 TAMs in NSCLC metastasis modulated RASSF4 to promote NSCLC progression.
Inhibiting miRNA-enriched exosomes from M2 TAMs is useful for treating cancer metastasis (53,54). In our work, we found that inhibiting exosomal miR-155 and miR-196a-5p could alleviate NSCLC metastasis activated by exosomes derived from M2 TAMs. Therefore, targeting exosomal miR-155 and miR-196a-5p from M2 TAMs may be a potential therapeutic strategy for NSCLC metastasis. Moreover, previous researches have also indicated that miRNA can control polarization of TAMs (55). Whether miR-155 and miR-196a-5p are involved in the M2 polarization of TAMs deserve further investigation, which is now underway in our group.
Overall, this study showed that M2 TAMs were the main population of TAMs in metastatic tissues of NSCLC and promoted NSCLC metastasis. The cross-talk between M2 TAMs and lung cancer cells was mediated by exosomes that carry miR-155 and miR-196a-5p. MiR-155 and miR-196a-5p, which were abundant in both M2 TAMs and exosomes from M2 TAMs, promoted EMT and migration of NSCLC through targeting RASSF4. Inhibition of miR-155 or miR-196a-5p in M2 TAMs could prevent NSCLC metastasis. These findings suggest an undescribed mechanism of exosomes transfer from M2 macrophages to NSCLC cells, which explains the way immune cells participate in tumor progression.
Acknowledgments
Funding: This work was supported in part by key medical research project of Jiangsu Provincial Health Commission (K2019002), Ethicon Excellence in Surgery Grant (HZB-20190528-13), and Jiangsu Province Natural Science Foundation (BK20201492).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at: http://dx.doi.org/10.21037/tlcr-20-1255
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-1255
Peer Review File: Available at http://dx.doi.org/10.21037/tlcr-20-1255
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at: http://dx.doi.org/10.21037/tlcr-20-1255). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The project has attained informed consent from all patients and was approved by the Ethics Committee of Jiangsu Province People’s Hospital and the First Affiliated Hospital of Nanjing Medical University (No. 2019-SR-266). The Laboratory Animal Care and Use Committees of the hospital approved all experimental procedures, which are in compliance with the national or institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Keith RL, Miller York E. Lung cancer chemoprevention: current status and future prospects. Nat Rev Clin Oncol 2013;10:334-43. [Crossref] [PubMed]
- Andrews J, Yeh P, Pao W, et al. Molecular predictors of response to chemotherapy in non-small cell lung cancer. Cancer J 2011;17:104-13. [Crossref] [PubMed]
- Nichols L, Rachel S, Friedrich K. Causes of death of patients with lung cancer. Arch Pathol Lab Med 2012;136:1552-7. [Crossref] [PubMed]
- Wang X, Adjei AA. Lung cancer and metastasis: new opportunities and challenges. Cancer Metastasis Rev 2015;34:169-71. [Crossref] [PubMed]
- Verma V, Lautenschlaeger T. MicroRNAs in non-small cell lung cancer invasion and metastasis: from the perspective of the radiation oncologist. Expert Rev Anticancer Ther 2016;16:767-74. [Crossref] [PubMed]
- Rosell R, Karachaliou N. Relationship between gene mutation and lung cancer metastasis. Cancer Metastasis Rev 2015;34:243-8. [Crossref] [PubMed]
- Popper HH. Progression and metastasis of lung cancer. Cancer Metastasis Rev 2016;35:75-91. [Crossref] [PubMed]
- Anderson NM, Simon M. BACH1 Orchestrates Lung Cancer Metastasis. Cell 2019;179:800. [Crossref] [PubMed]
- Wu SG, Chang TH, Liu YN, et al. MicroRNA in lung cancer metastasis. Cancers 2019;11:265. [Crossref] [PubMed]
- Pereira JL, Gomes M, Teixeira AL, et al. Potential and importance of metalloproteinases and interleukins in inflammation and metastasization in non-small cell lung cancer. Transl Cancer Res 2018;7:782-95. [Crossref]
- Spano D, Zollo M. Tumor microenvironment: a main actor in the metastasis process. Clin Exp Metastasis 2012;29:381-95. [Crossref] [PubMed]
- Wood SL, Pernemalm M, Crosbie PA, et al. The role of the tumor-microenvironment in lung cancer-metastasis and its relationship to potential therapeutic targets. Cancer Treat Rev 2014;40:558-66. [Crossref] [PubMed]
- Turley SJ, Cremasco V, Astarita Jillian L. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat Rev Immunol 2015;15:669-82. [Crossref] [PubMed]
- Nagarsheth N, Wicha MS, Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol 2017;17:559-72. [Crossref] [PubMed]
- De Palma M, Biziato D, Petrova TV. Microenvironmental regulation of tumour angiogenesis. Nat Rev Cancer 2017;17:457-74. [Crossref] [PubMed]
- Chanmee T, Ontong P, Konno K, et al. Tumor-Associated Macrophages as Major Players in the Tumor Microenvironment. Cancers 2014;6:1670-90. [Crossref] [PubMed]
- Lin Y, Xu J, Lan H. Tumor-associated macrophages in tumor metastasis: biological roles and clinical therapeutic applications. J Hematol Oncol 2019;12:76. [Crossref] [PubMed]
- Ma J, Liu L, Che G, et al. The M1 form of tumor-associated macrophages in non-small cell lung cancer is positively associated with survival time. BMC Cancer 2010;10:112. [Crossref] [PubMed]
- Yuan A, Hsiao YJ, Chen HY, et al. Opposite Effects of M1 and M2 Macrophage Subtypes on Lung Cancer Progression. Sci Rep 2015;5:14273. [Crossref] [PubMed]
- Guo Z, Song J, Hao J, et al. M2 macrophages promote NSCLC metastasis by upregulating CRYAB. Cell Death Dis 2019;10:377. [Crossref] [PubMed]
- Li Q, Li BW, Li Q, et al. Exosomal miR-21-5p derived from gastric cancer promotes peritoneal metastasis via mesothelial-to-mesenchymal transition. Cell Death Dis 2018;9:854. [Crossref] [PubMed]
- Zhang Y, Liu DQ, Chen X, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell 2010;39:133-44. [Crossref] [PubMed]
- Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654-9. [Crossref] [PubMed]
- Ying X, Wu QF, Wu XL, et al. Epithelial ovarian cancer-secreted exosomal miR-222-3p induces polarization of tumor-associated macrophages. Oncotarget 2016;7:43076-87. [Crossref] [PubMed]
- Yin Z, Ma TT, Huang BW, et al. Macrophage-derived exosomal microRNA-501-3p promotes progression of pancreatic ductal adenocarcinoma through the TGFBR3-mediated TGF-β signaling pathway. J Exp Clin Cancer Res 2019;38:310. [Crossref] [PubMed]
- Zhang M, Wang DP, Zhu TT, et al. RASSF4 Overexpression Inhibits the Proliferation, Invasion, EMT, and Wnt Signaling Pathway in Osteosarcoma Cells. Oncol Res 2017;25:83-91. [Crossref] [PubMed]
- Han Y, Dong Q, Hao J, et al. RASSF4 is downregulated in nonsmall cell lung cancer and inhibits cancer cell proliferation and invasion. Tumour Biol 2016;37:4865-71. [Crossref] [PubMed]
- Ma M, He M, Jiang Q, et al. MiR-487a Promotes TGF-β1-induced EMT, the Migration and Invasion of Breast Cancer Cells by Directly Targeting MAGI2. Int J Biol Sci 2016;12:397-408. [Crossref] [PubMed]
- Lobb RJ, Becker M, Shu WW, et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicles 2015;4:27031. [Crossref] [PubMed]
- Reck M, Heigener DF, Mok T, et al. Management of non-small-cell lung cancer: Recent developments. Lancet 2013;382:709-19. [Crossref] [PubMed]
- Kim YB, Ahn YH, Jung JH, et al. Programming of macrophages by UV-irradiated apoptotic cancer cells inhibits cancer progression and lung metastasis. Cell Mol Immunol 2019;16:851-67. [Crossref] [PubMed]
- Li H, Nan H, Weikang Z, et al. Modulation the crosstalk between tumor-associated macrophages and non-small cell lung cancer to inhibit tumor migration and invasion by ginsenoside Rh2. BMC Cancer 2018;18:579. [Crossref] [PubMed]
- Bardi GT, Smith MA, Hood JL. Melanoma exosomes promote mixed M1 and M2 macrophage polarization. Cytokine 2018;105:63-72. [Crossref] [PubMed]
- Goswami KK, Ghosh T, Ghosh S, et al. Tumor promoting role of anti-tumor macrophages in tumor microenvironment. Cell Immunol 2017;316:1-10. [Crossref] [PubMed]
- Sumitomo R, Hirai T, Fujita M, et al. M2 tumor-associated macrophages promote tumor progression in non-small-cell lung cancer. Exp Ther Med 2019;18:4490-8. [Crossref] [PubMed]
- Cianciaruso C, Beltraminelli T, Duval F, et al. Molecular Profiling and Functional Analysis of Macrophage-Derived Tumor Extracellular Vesicles. Cell Rep 2019;27:3062-80.e11. [Crossref] [PubMed]
- Zheng P, Luo Q, Wang W, et al. Tumor-associated macrophages-derived exosomes promote the migration of gastric cancer cells by transfer of functional Apolipoprotein E. Cell Death Dis 2018;9:434. [Crossref] [PubMed]
- Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem 2019;88:487-514. [Crossref] [PubMed]
- Bartel DP. Metazoan MicroRNAs. Cell 2018;173:20-51. [Crossref] [PubMed]
- Lan J, Sun L, Xu F, et al. M2 Macrophage-Derived Exosomes Promote Cell Migration and Invasion in Colon Cancer. Cancer Res 2019;79:146-58. [Crossref] [PubMed]
- Li J, Su TQ, Yang L, et al. High SLC17A9 expression correlates with poor survival in gastric carcinoma. Future Oncol 2019;15:4155-66. [Crossref] [PubMed]
- Li N, Cui T, Guo WL, et al. MiR-155-5p accelerates the metastasis of cervical cancer cell via targeting TP53INP1. Onco Targets Ther 2019;12:3181-96. [Crossref] [PubMed]
- Xu S, Shi L. High expression of miR-155 and miR-21 in the recurrence or metastasis of non-small cell lung cancer. Oncol Lett 2019;18:758-63. [Crossref] [PubMed]
- Xin H, Wang C, Liu Z. MiR-196a-5p promotes metastasis of colorectal cancer via targeting IκBα. BMC Cancer 2019;19:30. [Crossref] [PubMed]
- Pan J, Li X, Wu WJ, et al. Long non-coding RNA UCA1 promotes cisplatin/gemcitabine resistance through CREB modulating miR-196a-5p in bladder cancer cells. Cancer Lett 2016;382:64-76. [Crossref] [PubMed]
- Pan Y, Shu X, Sun LC, et al. miR-196a-5p modulates gastric cancer stem cell characteristics by targeting Smad4. Int J Oncol 2017;50:1965-76. [Crossref] [PubMed]
- Ni WJ, Wen JN. miRNA-dependent Activation of mRNA Translation. Microrna 2016;5:83-6. [Crossref] [PubMed]
- Li Z, Rana Tariq M. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov 2014;13:622-38. [Crossref] [PubMed]
- Donninger H, Schmidt ML, Mezzanotte J, et al. Ras signaling through RASSF proteins. Semin Cell Dev Biol 2016;58:86-95. [Crossref] [PubMed]
- Iwasa H, Hossain S, Hata Y. Tumor suppressor C-RASSF proteins. Cell Mol Life Sci 2018;75:1773-87. [Crossref] [PubMed]
- Michifuri Y, Hirohashi Y, Torigoe T, et al. Small proline-rich protein-1B is overexpressed in human oral squamous cell cancer stem-like cells and is related to their growth through activation of MAP kinase signal. Biochem Biophys Res Commun 2013;439:96-102. [Crossref] [PubMed]
- Fritz JM, Tennis MA, Orlicky DJ, et al. Depletion of tumor-associated macrophages slows the growth of chemically induced mouse lung adenocarcinomas. Front Immunol 2014;5:587. [Crossref] [PubMed]
- Chuang HY, Su YK, Liu HW, et al. Preclinical Evidence of STAT3 Inhibitor Pacritinib Overcoming Temozolomide Resistance via Downregulating miR-21-Enriched Exosomes from M2 Glioblastoma-Associated Macrophages. J Clin Med 2019;8:959. [Crossref] [PubMed]
- Essandoh K, Li Y, Huo J, et al. MiRNA-Mediated Macrophage Polarization and its Potential Role in the Regulation of Inflammatory Response. Shock 2016;46:122-31. [Crossref] [PubMed]


