
Plasma pre-treatment T790M relative allelic frequency in patients with advanced EGFR-mutated non-small cell lung cancer predicts treatment response to subsequent-line osimertinib
Introduction
Patients with epidermal growth factor receptor-mutated (EGFR+) non-small cell lung cancer (NSCLC) have a high response rate to first-line EGFR tyrosine kinase inhibitors (TKI) and typically have a better prognosis than EGFR-wildtype NSCLC patients. However, almost all patients will develop disease progression due to development of treatment resistance to first/second-generation EGFR-TKI after a median of 9 to 13 months on therapy (1-4). Approximately half of these patients will acquire a secondary EGFR mutation in exon 20 of EGFR, the T790M mutation. Substitution of methionine for threonine at position 790 (Thr790Met) in exon 20 of the EGFR confers acquired resistance to first/second-generation EGFR-TKI. The bulky methionine side chain causes steric hindrance, affecting the ability of first/second-generation EGFR-TKIs to bind to the ATP kinase pocket (5) and alters the affinity of EGFR to ATP as the favoured substrate (6). Detection of this secondary T790M mutation predicts treatment response to osimertinib (AZD9291), a third-generation EGFR-TKI, which targets both the original EGFR sensitising and T790M resistance mutations. Osimertinib was approved by the Therapeutic Goods Administration (TGA) in Australia in August 2016 (7), for the treatment of patients with locally advanced or metastatic EGFR T790M mutation positive NSCLC based on the AURA trials (8,9). Osimertinib has shown superior response rates and progression-free survival (PFS) compared with chemotherapy in this group of patients (median PFS 10.1 vs. 4.4 months; HR 0.3; 95% CI: 0.23–0.41; P<0.001) (9).
The National Comprehensive Cancer Network (NCCN) treatment guidelines advocate actionable mutation screening as standard of care (10), but sufficient tumour tissue to yield adequate DNA is often difficult to obtain, especially in the relapsed and metastatic settings (11). Clinically relevant mutations may also change during the course of treatment (12), an evolution that is difficult to monitor through sequential tissue biopsies. Spatial and temporal tumour heterogeneity makes accurate assessment of resistance mutations based on the biopsy of a single metastatic site challenging. A reliable and non-invasive method for detection of clinically actionable mutation is therefore essential for the effective delivery of precision medicine for patients with NSCLC.
Plasma circulating tumour DNA (ctDNA) testing to diagnose EGFR sensitising mutations and T790M mutation was approved by the FDA in 2015 (13). Patients with positive plasma testing showed similar response rates to osimertinib compared with patients who tested positive on tissue biopsy (14). Plasma testing for EGFR and T790M mutations can be done using several methods including polymerase chain reaction (PCR) and next generation sequencing (NGS) platforms. These methods will quantify the copy number of EGFR and T790M mutant and wildtype alleles. Currently, the NCCN guidelines also recommend the use of liquid biopsy as an alternative to tissue biopsy for initial T790M mutation testing. If the plasma biopsy is negative then tissue biopsy is recommended where feasible (15).
T790M relative allelic frequency (RAF) is defined as the proportion of T790M mutant allelic frequency (AF) compared with the EGFR driver mutant AF. This proportion varies from patient to patient due to underlying disease biology. Studies in tumour and cytology samples have shown that T790M AF might correlate with treatment response with osimertinib and survival outcome (16,17). In an exploratory analysis of the phase I dose escalation AURA study, Oxnard et al. (14) reported that increased T790M RAF was not associated overall with a greater depth of response (R =−0.183) to osimertinib, but patients with T790M RAF >10% showed a greater depth of response compared with those with T790M RAF <10%. To our knowledge, the impact of pre-treatment T790M resistance mutation RAF on disease control rate (DCR) and survival outcome has not been formally assessed in any large study using the standard full dose of osimertinib in the real-world setting. We hypothesise that as plasma testing could quantify mutation burden, baseline T790M mutation load could be a useful biomarker with prognostic value for patients with EGFR+ NSCLC.
Previous study using receiver operating characteristics (ROC) curve analysis has identified plasma T790M RAF cut-off values of 0.22 and 0.3 (18,19). These small studies showed that there is a difference in survival outcomes for patients with high vs. low T790M RAF at the respective cut-off points. Using the same T790M RAF cut-off point of 0.3, we report on the DCR and survival of patients treated with second/subsequent-line osimertinib stratified based on pre-treatment plasma T790M RAF. In addition, this study also investigates the DCR and survival of patients with tumour vs. plasma T790M positivity. We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-1125).
Methods
Patients
Eligible patients included those treated with osimertinib in the second-line setting and beyond. Prior to osimertinib being available on the Pharmaceutical Benefit Scheme (PBS) in Australia, patients with EGFR+ NSCLC who developed disease progression after first-line EGFR-TKI were eligible for T790M plasma testing in Australia with provision of osimertinib in those who tested positive (by plasma or tissue) via a compassionate access program provided by AstraZeneca between September 2016 and August 2018. From February 2019, osimertinib was reimbursed by the PBS in Australia based on tumour but not plasma T790M positivity. Therefore, we also included patients with tumour T790M positivity to determine if there is a difference in treatment outcome when compared with patients with plasma T790M positivity.
Data on demographics, disease characteristics, T790M RAF and treatment outcome were collected. All patients received osimertinib 80 mg daily as a starting dose, with dose reductions or interruptions according to the clinicians’ discretion due to treatment toxicity or disease progression. Plasma testing was conducted at three different national association of testing authorities (NATA) accredited laboratories (NATA is the authority that provides independent assurance of technical competence in Australia). All three laboratories used digital PCR platforms [BEAMing PCR (20,21) and droplet digital PCR (22)]. Only one laboratory used the exon 19 deletion assay that is designed to detect the loss of wild type sequence, which will be more capable of picking up the sequence deletion irrespective of the deletion variants {Seki, 2016 #297}. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by Human Research ethics committee (HREC) protocols from Royal Prince Alfred Hospital, Australia (Protocol number X18-0327, LNR/18/RPAH/462) and individual consent for this retrospective analysis was waived.
Efficacy assessment
Radiological treatment response data were retrieved from original radiological reports or documentation made by treating clinicians; there was no independent radiological imaging review. The first evaluation of treatment response was done at approximately 3 months of treatment with osimertinib and then assessed at approximately three to six monthly intervals until disease progression as per local standard practice. DCR was defined as complete response, partial response or stable disease. This was assessed at first radiological assessment at approximately 3 months of treatment.
Overall survival (OS) was defined as the time from the first dose of osimertinib to the date of death due to any cause. Patients who were alive at the time of the final analysis or who became lost to follow-up were censored at their last known alive date. Progression-free survival (PFS) was defined as the time from the first dose of osimertinib to the date of the first documented disease progression or death. Patients who remained progression-free and still alive at the time of analysis were censored on the date of their last clinical follow-up.
Data was collected by reviewing patients’ electronic or paper medical records; with missing data, attempts were made to retrieve this data by consulting individual clinicians.
Determination of allelic frequency (AF) and T790M relative allelic frequency (RAF)
AF of each somatic mutation was calculated as the percentage of mutant DNA allele reads relative to total DNA allele reads (mutant plus wildtype). The T790M RAF was calculated as the ratio of T790M allelic frequency to EGFR driver allelic frequency. In patients where EGFR driver mutation was not detected (n=10), we determined this was likely due to the low sensitivity of EGFR activating mutation assays to detect low AF in these samples, therefore the AF for EGFR activating mutations will be lower than AF for T790M mutation for these samples. We categorised the RAF ratio to be >0.3.
Statistical analysis
Patient demographics, disease and treatment information were analysed descriptively (Table 1). PFS and OS were estimated using Kaplan-Meier analysis. Cox regression was used to determine whether relative allelic frequency (RAF) of T790M was associated with OS and PFS. Fisher’s exact test was used to determine whether RAF is associated with DCR. All analyses were performed using IBM SPSS Statistics (Version 25) and GraphPad Prism (Version 8.4.3) and P values were calculated based on deviation from zero with P<0.05 being statistically significant. Graphs were generated in GraphPad Prism 8.4.3.
Full table
The primary objective was to investigate if there was a difference in DCR between patients with high vs. low relative allelic frequency (RAF) assessed from ctDNA, using a 0.3 RAF cut-off. Secondary objectives include survival outcome in patients with high vs. low RAF, and patients with plasma versus tissue T790M positivity.
Results
Patients
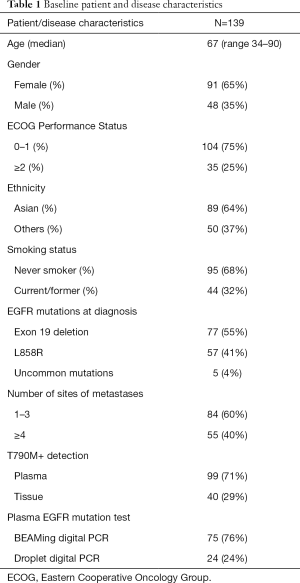
A total of 147 patients were identified from ten different sites in Australia (Table 1). Eight patients were excluded due to the reasons listed in Figure 1. Of the 139 included patients, the majority were female (65%), most had Eastern Cooperative Oncology Group (ECOG) performance status of ≤1 (75%), 64% were of Asian ethnicity, and 68% were never smokers. Five patients (4%) had uncommon EGFR mutations detected at baseline (2 with compound mutations, 1 with G719S, 1 with L861Q, 1 with S768I). 80% of patients received osimertinib as second-line therapy and 20% received osimertinib as third/fourth-line therapy. Table 1 summarises the demographic and disease characteristics of the patients.
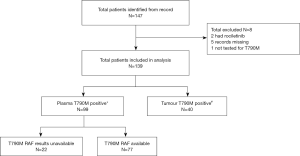
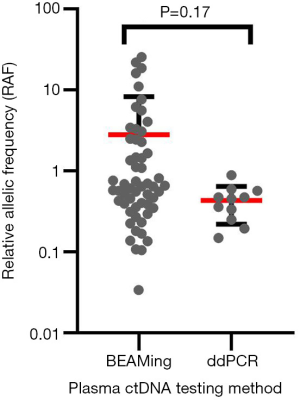
Ninety-nine patients (71%) were treated based on plasma T790M positivity and 40 (29%) based on tissue T790M positivity. Eleven/99 patients who were plasma T790M+ tested negative on tissue for T790M. Of the 40 patients who were tissue T790M+, 25 patients had an upfront tissue biopsy with no plasma testing and 15 had tissue biopsy due to negative plasma T790M results. In total, 26 patients had both tumour and plasma T790M results available. For this real-world study, a tissue test was only performed if liquid biopsy was negative (or vice versa). Therefore, patients who were tested EGFR T790M positive on a second test always had a discordant result (Figure 1). All plasma testing was done using digital PCR platforms (84% BEAMing, 16% droplet digital PCR) (Figure 2).
Overall, DCR was 73%, median progression-free survival (PFS) was 12.3 months (96% CI: 9.7–14.9 months) and median overall survival (OS) was not reached. Median follow-up duration was 17.1 months for PFS and OS analyses.
High vs. low pre-treatment plasma T790M RAF
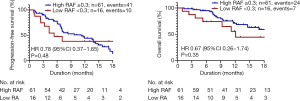
The median plasma pre-treatment T790M RAF for this patient cohort (n=77) was 0.64. Using pre-determined RAF cut-off of 0.3, patients with higher RAF (≥0.3) showed higher DCR compared with those in the lower RAF (<0.3) group (72% vs. 36%, P=0.02). The median PFS for patients with higher RAF was numerically longer compared with patients with lower RAF (median PFS 11.2 vs. 5.4 months; median OS not reached), but this was not statistically significant (Figure 3).
Plasma vs. tissue T790M positivity
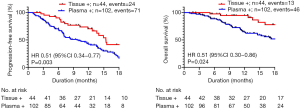
Exploratory analysis showed that patients with tissue T790M+ had better disease control rates compared with those with plasma T790M+ (89% vs. 68%, P=0.01). Patients with tissue T790M+ also had longer PFS (median 15.4 vs. 9.7 months; HR 2.12, 95% CI: 1.38–3.25, P=0.02) and OS (median not reached; HR 2.65, 95% CI: 1.49–4.74, P=0.008) compared to those who were plasma positive (Figure 4). 70% of patients who were tissue T790M+ had lower metastatic disease burden (≤3 sites of metastatic disease) compared with 58% of patients who were plasma T790M+ for our patient cohort. However, there was no statistical association between tissue/plasma T790M positivity and disease burden (P=0.25).
Discordant plasma vs. tissue T790M status
A small proportion of patients had discordant tumour/plasma T790M positivity. Of the 13 patients who were plasma T790M− and had tissue biopsy with tumour T790M+, DCR was 77%; of the 11 patients who had upfront tissue biopsy with tumour T790M− and subsequently was plasma T790M+, DCR was 64%.
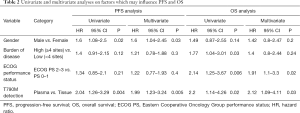
Univariate and multivariate analysis
We performed univariate and multivariate analyses on factors which may influence PFS or OS (Table 2). The variables included in the multivariate model were those that were found to be associated with survival on univariate analyses or previously reported to be associated with survival in clinical trials. Gender, burden of disease, ECOG performance status and method of T790M detection (plasma vs. tissue) were chosen as the variables for the multivariate model. Other clinical factors which may influence survival such as smoking history, age and brain metastases were not included as they were not associated with survival on univariate analysis. Patients who were tissue T790M+ maintained a superior survival (PFS and OS) compared to plasma T790M+ after accounting for other prognostic variables.
Full table
Discussion
To our knowledge, this is the first multi-centre study based on real-world clinical data that shows a relationship between pre-treatment specific plasma mutation burden with disease outcomes for patients treated with therapy targeted at a specific mutation. We showed that higher levels of T790M RAF in plasma, is a predictor of disease control in patients with T790M+ NSCLC treated with osimertinib. In addition, we found that patients who were tissue T790M+ had better tumour response (89% vs. 68%) and survival (median PFS difference of 5.7 months) compared with patients who were plasma T790M+ when being treated with osimertinib. However, the DCR was still high at 68% in patients with plasma T790M+ disease. Superior response to osimertinib for tissue T790M+ remained in a multivariate model adjusting for other confounding variables.
Multiple different resistance mechanisms can develop within the same tumour and at different metastatic sites within the same patient (23,24). Therefore, when a patient develops acquired resistance after treatment with a first- or second-generation EGFR TKI, in addition to T790M+ clones, T790M− clones harbouring EGFR-dependent or EGFR-independent resistant mechanisms may also occur concurrently (25). As osimertinib is only effective for tumour clones harbouring EGFR sensitising or T790M mutations, investigating surrogates that can represent the proportion of T790M+ cells and predict how effective osimertinib will be in each patient is essential. There are currently prospective studies investigating if alternating first-generation EGFR-TKI gefitinib with osimertinib in the first line setting could affect the appearance of the T790M+ and T790M− clones and therefore affect treatment outcome, i.e., OSCILLATE study, ACTRN 12617000720314 and APPLE study, NCT02856893 (26).
Digital PCR platforms such as BEAMing and ddPCR used for plasma mutation testing for this study are both highly sensitive and specific. These non-invasive methods detect and quantify mutant and wildtype alleles in plasma and are now arguably more widely available and could be integrated into diagnostic testing at a lower cost compared to NGS. There has been much interest in the analysis of surrogates, such as T790M and EGFR sensitising mutation allelic frequency (AF) to predict response to osimertinib with inconsistent results observed (18,27,28). In our study, we have chosen T790M RAF as this value accounts for both T790M and EGFR sensitising mutation AF, which is a better surrogate for the proportion of tumour clones that are T790M+. T790M RAF could be obtained easily from the results analysed by digital PCR platforms for each patient. We used a pre-determined T790M RAF threshold of 0.3 based on previous studies that dichotomise T790M using ROC curve analysis (18,19). We believe that by having a set threshold for T790M RAF rather than having it as a continuous variable will simplify risk stratification in the clinic and allow clinicians to use this surrogate marker in clinical practice.
Tumour heterogeneity at disease progression with first-line EGFR-TKI was previously reported and studies have shown that a high tumour proportion for T790M clones in tumour biopsy predicts a better response to third generation T790M targeting EGFR-TKIs rociletinib (16) and osimertinib (17,29). Plasma T790M RAF has also been shown to correlate with treatment response to osimertinib (14,17,18). To our knowledge, this is the first large study using real-world data which showed pre-treatment T790M RAF in plasma could be used as a predictive biomarker to osimertinib response. However, despite having better disease control rate to osimertinib, we did not find a significant difference in PFS and OS for the group with high vs. low T790M RAF. The number of patients with low T790M RAF is relatively small (n=16) with relatively low event rates, i.e., 10/16 (63%) for PFS analysis and 7/16 (44%) for OS analysis (Figure 3). Therefore small subgroup size could have contributed to the absence of statistically significant differences for survival analyses.
The second part of our study has shown that patients who were tissue T790M+ had better disease control and survival when treated with osimertinib compared with the group who was plasma T790M+. This finding is different to another published work where patients had similar response rates and survival outcome whether they were treated with osimertinib based on plasma or tissue T790M positivity (14). However, Oxnard et al. used patient data from larger prospective clinical trials (AURA phase 1 escalation and expansion cohorts), with a different patient population due to strict trial selection criteria. Almost all patients for the aforementioned study had both tissue and plasma testing done, and with approximately 90% of patients who were considered plasma T790M+ were also tissue T790M+. In contrast, most patients included in our real-world study only had one (tissue or plasma) test done. From that perspective, it is possible that a higher proportion of patients in our study were plasma T790M+ but tissue T790M−.
Less favourable clinical outcomes in patients who were plasma T790M+ could be partially related to the possibility that these patients, with detectable tumour DNA in the plasma, have larger tumour size, disease burden and extra-thoracic disease (30,31). This is consistent with our previous study, in which EGFR+ NSCLC patients with detectable ctDNA at baseline had higher burden of metastatic disease (32). Further, as resistant cancers are inherently more heterogeneous than treatment-naive cancer, highly sensitive plasma testing could detect small amounts of T790M minor clones which may not be detected with a single tumour biopsy. Some of these minor clones of T790M may not be the main mechanism of resistance, and therefore may partly explain the unfavourable outcome of patients who were plasma T790M+ on osimertinib.
Even though plasma mutation testing and ctDNA sequencing are now more widely available with impressive data for their use to detect T790M mutation, they are still not in routine use in clinical practice where tissue-based molecular testing remains the standard of care. In Australia, plasma ctDNA testing for EGFR or T790M mutations does not attract a Medicare rebate with significant out-of-pocket charge to patients. In addition, patients who are plasma T790M+ but tissue T790M unknown or negative will not be able to access osimertinib under the Australia PBS scheme.
Although tissue biopsy testing has the advantage of allowing the detection of changes in histology (e.g., small cell transformation) and assessment of other mutational changes causing resistance, the heterogeneity within the tumour and rates of T790M positivity at different metastatic sites can vary widely (7). Plasma ctDNA mutation testing and sequencing can provide a more representative profile of the overall predominant resistance mechanisms for a given patient than a core biopsy from one region of a single metastatic lesion. This study supports the advantage of liquid biopsy, over tissue biopsy sampling, which could provide quantitative information (i.e., RAF) from multiple tumour sites which could be used as a predictive marker in the management of EGFR+ NSCLC patients treated with osimertinib. A small proportion of patients (n=11) had negative tumour biopsy but were plasma T790M positive, this group of patients still showed high DCR of 64% in our study which once again support the utility of plasma mutation testing for advanced EGFR+ NSCLC patients where tumour biopsy is not possible. In common with other studies, our data suggest that those patients with tumour T790M positivity had better outcomes with osimertinib.
The extent of tumour heterogeneity at treatment resistance to first-line EGFR-TKI with variable degrees of T790M+ clones could explain why some patients do not respond to third-generation EGFR-TKI osimertinib despite being T790M+ positive in tissue or plasma. The clinical implication of this is that combination regimens that also target non-T790M clones are required as acquired resistance mechanisms do not happen in a binary (positive/negative) fashion. Due to molecular heterogeneity, some T790M ‘positive’ cancers have distinct resistant clones besides T790M.This study supports the growing lines of evidence that treatment strategies that combine osimertinib with other agents that target T790 wildtype cells will be more likely to produce more durable responses.
We acknowledge that there are limitations for this study. Firstly, plasma EGFR sensitising and T790M mutation testing were performed in three separate labs in Australia using two different PCR platforms for analysis of RAF for our patient cohort. BEAMing digital PCR and ddPCR were used which have different accuracies at detecting EGFR sensitising and T790M mutations. However, we believe that the difference will be minimal as a cross-platform comparison study showed that both these platforms have the same sensitivity at detecting T790M (71%) with similar concordance rate (70–74%); albeit with ddPCR showing slightly higher specificity compared with BEAMing platform (83% vs. 67%) (33). We also noted that there was a significant proportion of patient samples with T790M RAF >1 using BEAMing assay. This could be related to the suboptimal sensitivity or specificity of T790M or EGFR sensitising mutation assays. However, this accurately reflects the data used for determining patient eligibility for treatment at this point in time and is included as provided by the NATA accredited laboratory. In addition, not all laboratories used the most optimal method for plasma EGFR exon 19 deletion detection that was designed to detect the loss of wildtype sequence, which may lead to false negativity. Unfortunately, the pre-analytical aspects of each testing platforms are beyond the scope of this retrospective analysis and we acknowledge that this is a limitation of this study. Secondly, despite the multivariate analysis showing tissue T790M+ as an independent prognostic marker, other potential confounders that could affect the survival outcome for plasma vs. tissue T790M+ were not available, such as tumour size (34), discordant plasma and tissue T790M status (14) and presence of extra-thoracic disease (35). Finally, the limited sample size and the retrospective design of this study may yield statistical bias and undefined biases may have existed and influenced clinical outcomes. Further large prospective multi-centre validation trials with uniform testing facilities and PCR platform is needed to confirm our findings. Our group has previously shown that clearance of plasma ctDNA for EGFR mutations is associated with better treatment outcome; with a rise in plasma ctDNA predicting treatment failure (32). For this study, patients only had one-off plasma testing, a follow-up study is warranted to elucidate temporal changes in T790M RAF and how this correlates with treatment outcome.
In conclusion, this study shows that high pre-treatment T790M RAF ≥0.3 is associated with superior disease control for advanced EGFR+ T790M+ NSCLC patients treated with osimertinib. Our findings also suggest that tissue T790M positivity may be an independent prognostic marker for this group of patients over plasma T790M positivity. These findings require confirmation in prospective trials.
Acknowledgments
Presented in part at the Plenary Session of the Medical Oncology Group Australia (MOGA) annual scientific meeting, Canberra, Australia, August 14-16, 2019. We would like to thank all the patients and their families who are involved in this study. We are grateful for all the clinicians involved at the individual sites who has assisted with site-specific ethics application and data collection. We would also like to thank CONCERT Biobank team for helping with HREC submission. We thank Dr. Sonia Yip for her help in identifying eligible patients from the LC-PLAT lung cancer cohort. We are grateful for the statistical advice from Dr. Wei Xuan, senior biostatistician at The Ingham Institute.
Funding: This work was supported by an independent research grants received from AstraZeneca (ESR-17-12953) and the Cancer Institute NSW (CINSW) funded Centre for Oncology Education and Research Translation (CONCERT). PD is funded by an Australian Postgraduate Scholarship through Western Sydney University and by a top-up scholarship from CONCERT. TB is also funded through CONCERT. TR has been supported by a CINSW FRL fellowship and is a current Irene and Arnold Vitocco Cancer Research Fellow.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-1125
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-1125
Peer Review File: Available at http://dx.doi.org/10.21037/tlcr-20-1125
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-1125). Dr. Ding reports grants from Astrazeneca, during the conduct of the study; personal fees from Roche, personal fees from Astrazeneca, outside the submitted work; Dr. Lee reports grants and personal fees from Astrazeneca, personal fees from Boehringer Ingelheim, personal fees from Norvatis, personal fees from Pfizer, personal fees from Roche, outside the submitted work; Dr. Itchins reports personal fees from Astrazeneca, personal fees from Roche, personal fees from MSD, personal fees from Pfizer, personal fees from Takeda, personal fees from Novartis, outside the submitted work; Dr. Clarke reports personal fees from Astrazeneca, outside the submitted work; Dr. Blinman reports personal fees from Astrazeneca, personal fees from Boehringer Ingelheim, outside the submitted work; Dr. Kao reports personal fees from Astrazeneca, personal fees from Boehringer Ingelheim, personal fees from Pfizer, personal fees from MSD, personal fees from BMS, personal fees from Takeda, personal fees from Roche, outside the submitted work; Dr. John reports personal fees from Roche, personal fees from BMS, personal fees from Merck, personal fees from Ignyta, personal fees from Astrazeneca, personal fees from Takeda, personal fees from MSD, personal fees from Specialised therapeutics, personal fees from Pfizer, outside the submitted work; Dr. Leal reports grants and personal fees from Roche, grants and personal fees from BMS, grants and personal fees from Astrazeneca, grants and personal fees from MSD, grants and personal fees from Pfizer, grants and personal fees from Boehringher Ingelheim, personal fees from Sanofi, personal fees from Elly-Lilly, personal fees from Tecnofarma, outside the submitted work. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by Human Research ethics committee (HREC) protocols from Royal Prince Alfred Hospital, Australia (Protocol number X18-0327, LNR/18/RPAH/462) and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or Chemotherapy for Non–Small-Cell Lung Cancer with Mutated EGFR. N Engl J Med 2010;362:2380-8. [Crossref] [PubMed]
- Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010;11:121-8. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12:735-42. [Crossref] [PubMed]
- Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005;352:786-92. [Crossref] [PubMed]
- Yun CH, Mengwasser KE, Toms AV, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A 2008;105:2070-5. [Crossref] [PubMed]
- John T, Bowden JJ, Clarke S, et al. Australian recommendations for EGFR T790M testing in advanced non–small cell lung cancer. Asia Pac J Clin Oncol 2017;13:296-303. [Crossref] [PubMed]
- Janne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015;372:1689-99. [Crossref] [PubMed]
- Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Ettinger DS, Wood DE, Akerley W, et al. NCCN Guidelines Insights: Non–Small Cell Lung Cancer, Version 4.2016. J Natl Compr Canc Netw 2016;14:255-64. [Crossref] [PubMed]
- Sholl LM, Aisner DL, Varella-Garcia M, et al. Multi-institutional Oncogenic Driver Mutation Analysis in Lung Adenocarcinoma: The Lung Cancer Mutation Consortium Experience. J Thorac Oncol 2015;10:768-77. [Crossref] [PubMed]
- Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-92. [Crossref] [PubMed]
- U.S. Food and Drug Administration: Medical Devices: cobas EGFR Mutation Test v2 - P150047. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpma/pma.cfm?id=P150047
- Oxnard GR, Thress KS, Alden RS, et al. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3375-82. [Crossref] [PubMed]
- NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines). Non-small cell lung cancer. Version 3.2020. Available online: www.nccn.org.
- Piotrowska Z, Niederst MJ, Karlovich CA, et al. Heterogeneity Underlies the Emergence of EGFRT790 Wild-Type Clones Following Treatment of T790M-Positive Cancers with a Third-Generation EGFR Inhibitor. Cancer Discov 2015;5:713-22. [Crossref] [PubMed]
- Ariyasu R, Nishikawa S, Uchibori K, et al. High ratio of T790M to EGFR activating mutations correlate with the osimertinib response in non-small-cell lung cancer. Lung Cancer 2018;117:1-6. [Crossref] [PubMed]
- Del Re M, Bordi P, Rofi E, et al. The amount of activating EGFR mutations in circulating cell-free DNA is a marker to monitor osimertinib response. Br J Cancer 2018;119:1252-8. [Crossref] [PubMed]
- Zheng Q, Hong S, Huang Y, et al. EGFR T790M relative mutation purity predicts osimertinib treatment efficacy in non-small cell lung cancer patients. Clin Transl Med 2020;9:17. [Crossref] [PubMed]
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985-90. [Crossref] [PubMed]
- Garcia-Foncillas J, Alba E, Aranda E, et al. Incorporating BEAMing technology as a liquid biopsy into clinical practice for the management of colorectal cancer patients: an expert taskforce review. Ann Oncol 2017;28:2943-9. [Crossref] [PubMed]
- Hindson BJ, Ness KD, Masquelier DA, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 2011;83:8604-10. [Crossref] [PubMed]
- Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240-7. [Crossref] [PubMed]
- Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A 2007;104:20932-7. [Crossref] [PubMed]
- Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016;7:11815. [Crossref] [PubMed]
- Remon J, Menis J, Hasan B, et al. The APPLE Trial: Feasibility and Activity of AZD9291 (Osimertinib) Treatment on Positive PLasma T790M in EGFR-mutant NSCLC Patients. EORTC 1613. Clin Lung Cancer 2017;18:583-8. [Crossref] [PubMed]
- Buder A, Hochmair MJ, Schwab S, et al. Cell-Free Plasma DNA-Guided Treatment With Osimertinib in Patients With Advanced EGFR-Mutated NSCLC. J Thorac Oncol 2018;13:821-30. [Crossref] [PubMed]
- Helman E, Nguyen M, Karlovich CA, et al. Cell-Free DNA Next-Generation Sequencing Prediction of Response and Resistance to Third-Generation EGFR Inhibitor. Clin Lung Cancer 2018;19:518-30.e7. [Crossref] [PubMed]
- Kuo CS, Huang CH, Liu CY, et al. Prior EGFR-TKI Treatment in EGFR-Mutated NSCLC Affects the Allele Frequency Fraction of Acquired T790M and the Subsequent Efficacy of Osimertinib. Target Oncol 2019;14:433-40. [Crossref] [PubMed]
- Papadimitrakopoulou VA, Han JY, Ahn MJ, et al. Epidermal growth factor receptor mutation analysis in tissue and plasma from the AURA3 trial: Osimertinib versus platinum-pemetrexed for T790M mutation-positive advanced non-small cell lung cancer. Cancer 2020;126:373-80. [Crossref] [PubMed]
- Kwok WC, Ho JCM, Lam DCL, et al. When compared to plasma-based detection, osimertinib-treated non-small cell lung cancer (NSCLC) with tissue rebiopsy-confirmed acquired T790M mutation is associated with better survival. Asia Pac J Clin Oncol 2021;17:e35-9. [Crossref] [PubMed]
- Ding PN, Becker TM, Bray VJ, et al. The predictive and prognostic significance of liquid biopsy in advanced epidermal growth factor receptor-mutated non-small cell lung cancer: A prospective study. Lung Cancer 2019;134:187-93. [Crossref] [PubMed]
- Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015;90:509-15. [Crossref] [PubMed]
- Zhang J, Gold KA, Lin HY, et al. Relationship between tumor size and survival in non-small-cell lung cancer (NSCLC): an analysis of the surveillance, epidemiology, and end results (SEER) registry. J Thorac Oncol 2015;10:682-90. [Crossref] [PubMed]
- Lin JJ, Cardarella S, Lydon CA, et al. Five-Year Survival in EGFR-Mutant Metastatic Lung Adenocarcinoma Treated with EGFR-TKIs. J Thorac Oncol 2016;11:556-65. [Crossref] [PubMed]


