
Distinct patterns of somatic genomic alterations and mutational signatures in central and peripheral-type small-cell lung cancer
Introduction
Small-cell lung cancer (SCLC) is a highly malignant and widely metastatic subtype of lung cancer, which accounts for approximately 15% of lung cancer cases (1,2). SCLC is characterized by rapid tumor proliferation, genomic instability and mutation, high angiogenesis, and early metastasis (2,3). SCLC usually originates in the bronchi and is caused by smoking. Most patients with SCLC are current or former heavy smokers. Smoking is associated with increased mutation burdens of multiple distinct mutational signatures, which contribute to the development of SCLC (4-6).
In the past, some studies have done key research on the genetic landscape of SCLC by whole-genome or whole-exome sequencing (WES) (4,5,7,8). These studies confirmed the nearly universal inactivation of TP53 and RB1 in SCLC. Defective function of tumor suppressor genes TP53 and RB1 is known as early events in SCLC, which can be induced by mutation, loss of heterozygosity, or hemizygous deletion (DEL). Furthermore, compared with most cancers, higher genomic instability in SCLC was known, with an average tumor mutation burden (TMB) of 5.5 to 7.4 mutations per megabase, and about 175 mutations per tumor (9-11). Moreover, G-to-T transversions were frequently found in SCLCs, which is a tobacco carcinogenesis signature. There were also momentous structural alterations in SCLCs, such as DELs of 3p, 13q (which includes RB1), and 17p (which includes TP53), as well as amplifications (AMPs) of 5p and 3q (9,11). Although these studies characterized the genomic landscape of alterations in SCLCs, the details of the subtypes of SCLCs remain unclear.
SCLC can be subgrouped into peripheral- and central-type according to the location of the primary tumor (3). Primary tumors involving segmental or more proximal bronchi were defined as central-type SCLCs. Primary tumors involving subsegmental or more distal bronchi were defined as peripheral-type SCLCs. Only a few studies have mentioned the differences in survival between patients with central and peripheral-type SCLCs (12-14). However, research is scarce into genomic landscape and genetic heterogeneity between different origin positions in SCLCs, especially central and peripheral SCLCs.
In our study, a total of 41 tumor/control pairs passed the quality control (QC) were to move forward for WES. The average depth of the tumor DNA sequencing was 484×, and the control was 229×. We identified 8,186 genes with non-synonymous mutations in all cases, and analyzed distinct patterns of somatic genomic alterations and mutational signatures in central and peripheral-type SCLCs. Our findings uncovered that somatic genome alterations in central and peripheral-type SCLC might have different effects on tumorigenesis, immunotherapy, and prognosis. We present the following article in accordance with the MADR reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-1096).
Methods
Patients
Forty-one SCLC patients who were diagnosed in Shanghai Pulmonary Hospital from Nov 2018 to Jun 2019 were enrolled into this study. We did biopsy before any anti-cancer treatment. Experimental plans and protocols for this study (No. K18-203Y) were approved by the ethics/licensing committee of the Shanghai Pulmonary Hospital for review and confirmation. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from all patients participating in the study.
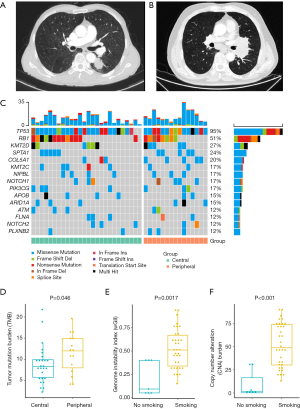
All patients underwent multidetector CT imaging (1-mm section thickness) at the time of diagnosis and the location of primary tumor (central or peripheral) was evaluated by experienced diagnostic experts (LW and JW) and, in the case of split opinions, other physicians (YH and CZ) were also involved in discussion with careful distinction of primary and metastatic tumors, leading to consensus. Based on previous reports (3,13), tumors involving segmental or more proximal bronchi were defined as a central-type, whereas tumors involving subsegmental or more distal bronchi were defined as a peripheral-type. Typical peripheral and central-type SCLCs are shown in Figure 1A,B.
Sample preparation, next-generation sequencing and data processing for WES
For the biopsy samples, DNA was extracted using the QIAamp DNA tissue kit (QIAGEN, Valencia, CA, USA) following the manufacturer’s instructions. For blood samples, 10 mL blood were collected in tubes containing EDTA and centrifuged at 1,600 ×g for 10 min at 4 °C within 2 h of collection. The peripheral blood lymphocyte (PBL) debris was stored at –20 °C until further use. The supernatants were further centrifuged at 10,000 ×g for 10 min at 4 °C, and plasma was harvested and stored at –80 °C until further use. DNA from PBLs was extracted using the RelaxGene Blood DNA system (Tiangen Biotech Co., Ltd., Beijing, China). Both cancer tissue and white blood cell genomic DNA was quantified with the Qubit 2.0 Fluorometer and the Qubit dsDNA HS assay kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to manufacturer’s instructions. In brief, fragmented genomic DNA underwent end-repairing, A-tailing and ligation with indexed adapters sequentially, followed by size selection using Agencourt AMPure XP beads (Beckman Coulter Inc., Brea, CA, USA), and DNA fragments were used for library construction using the KAPA Library Preparation kit (Kapa Biosystems, Inc., Wilmington, MA, USA) according to the manufacturer’s protocol. Hybridization-based target enrichment was carried out with HaploX WESPlus gene panel (an upgraded version of the standard WES, HaploX Biotechnology) for cancer tissue sequencing. Pre-LM-PCR Oligos (Kapa Biosystems, Inc.) in 50 µL reactions, and 7–8 PCR cycles were performed depending on the amount of DNA input. DNA sequencing was then performed on the Illumina Novaseq 6000 system. The average depth of the tumor DNA sequencing was 484×, and the control was 229×.
Somatic mutations, copy number alteration (CNA), and tumor’s clonal composition detection
Sequencing data were aligned to the UCSC hg19 genome using Burrows-Wheeler Aligner (BWA) (15). The BAMs were sorted and removed PCR duplications with sambamba (16). Base Quality Score Recalibration (BQSR) was performed with GATK 4.0 (17). Somatic mutation calling was performed using Strelka2 (18). Sequencing depth in each somatic mutation region as required to be ≥20 and ≥5 sequence reads had to support the variant call. In contrast, the number of reads supporting the variant in the germline data had to be <5 and sequencing depth was also required to be ≥20. Somatic CNAs (SCNAs), purity or ploidy was evaluated by FACETS (19).
Variant annotation
The variant data were annotated using ANNOVAR (20). Then common germline mutations with populational frequencies ≥1% were filtered out, using gnomAD, ExAC and esp6500 database. Variants were converted to MAF files using vcf2maf (https://github.com/mskcc/vcf2maf). Oncogenic pathway analysis was performed according to a previous work (21).
TMB
TMB was defined as the number of somatic coding mutations, including single base substitution and indels, per megabase of genome examined. Only nonsynonymous mutations were considered, excluding intron, synonymous mutations that do not affect the amino acid sequences. In detail, mutations were selected followed the instructions of maftools (22), which belongs to these ENSEMBL’s items: “frame shift del”, “frame shift ins”, “splice site”, “translation start site”, “nonsense mutation”, “nonstop mutation”, “in frame del”, “in frame ins”, and “missense mutation”.
Significantly mutated genes (SMGs) and frequently occurring SCNAs
SMGs were examined using MutSigCV (23). Both central and peripheral-type SCLC mutation data were preprocessed with maftools, and underwent MutSigCV analysis, using the default parameters. Frequently occurring SCNAs of central and peripheral-type SCLC were studied by GISTIC2 (24) independently, with the same parameters as the TCGA project’s pipeline (https://docs.gdc.cancer.gov/Data/Bioinformatics_Pipelines/CNV_Pipeline/).
Weighted genome integrity index, CNA burden, and intratumor heterogeneity (ITH)
The weighted genome instability index (wGII) was calculated as a previous work (25). Briefly, the integer ploidy number of each tumor sample was assessed by FACETS. The percentage of chromosomal regions with copy number gained or lost relative to the ploidy of the sample was calculated for every 22 autosomal chromosomes. Then the mean value of the 22 percentage values was determined as the wGII score. CNA burden was also estimated in a manner similar to Wolf et al. (26). In order to investigate the clonal and subclonal composition of the tumors, the somatic single nucleotide variants, SCNAs, and tumor purity data were used to perform clustering with PyClone (27). Somatic indels were excluded, and chromosomal regions were assessed as 0 copy numbers by FACETS were also excluded. Clusters with only one mutation were removed as noises of clustering. The number of clusters generated by PyClone was recorded as ITH index, and Shannon diversity index (SDI) was calculated as previously described (26).
Mutational signature analysis
Mutational Signatures were investigated using the MutationalPatterns R package (28). De novo mutational signature extraction was performed using the NMF algorithm (29). Cosine similarities between de novo extracted signatures and the COSMIC cancer mutational signatures were also calculated with MutationalPatterns. Absolute and relative contributions of each de novo obtained signature were also determined.
Statistical analysis
Most of the statistic was performed in R and ggpubr package. Fisher’s exact test or a non-parametric test was employed to correlate clinical and biological variables when necessary. The survival data were analyzed using the Kaplan-Meier (K-M) method, and differences in variables were compared using log-rank tests. Data visualizations were performed with maftools, ggpubr (30), and survminer (https://github.com/kassambara/survminer).
Results
Genomic landscape of Chinese SCLC and correlations between SMG, TMB, CIN, CNA burden and clinical characteristics
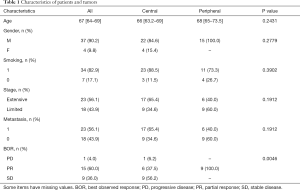
Base on patient CT-imaging and bronchoscopy results, we collected pathological specimens of 26 cases of classic central-type SCLCs and 15 samples of classic peripheral-type SCLCs. A total of 41 tumor/control pairs passed the QC were to move forward for WES, and we identified 8,186 genes with non-synonymous mutations in all cases. Clinical characteristics of all the patients are summarized in Table 1. There was little difference between the two groups except for best observed response (BOR). The BOR data are seriously missing, which may be the reason for the difference. Tumor metastasis occurred in 56.1% of the patients. The location of metastasis is very complex, including lung, pericardial effusion, liver, adrenal gland, thoracic spine, pleural effusion, pleura, brain, retroperitoneal lymph nodes, bone and other parts, as well as combinations of these positions. Thus, we analyzed the merged state of metastasis. In addition, none of the patients received surgical treatment. As shown in Figure 1B, TP53 and RB1 were the top 2 driver genes in both central and peripheral-type SCLCs. TP53 occurred in 100% of peripheral cases, and 92% of central cases. The occurring rates of RB1 were 53% and 50% respectively. MutSigCV analysis revealed two SMGs with adjusted P values <0.01 in both central and peripheral-type SCLCs respectively, which were TP53 and RB1 (Figure 1C and Table S1). These discoveries suggested that TP53 and RB1 were almost universally driver genes in central and peripheral-type SCLCs.
Full table
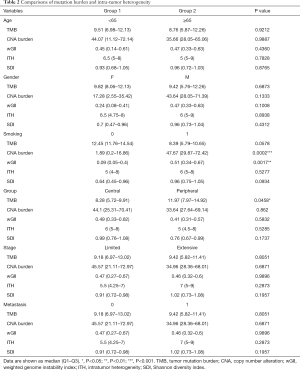
According to the hypothesis that tumors with higher TMB present more neoantigens and are more immunogenic (26), we examined the correlations between TMB and clinical characteristics such as group (central or peripheral-type), metastasis, age, gender, and smoking. As shown in Table 2, the median TMB of central-type SCLCs was 8.28 mutations per megabase (mut/Mb), by contrast, the median TMB of peripheral-type SCLCs was 11.97 mut/Mb. Compare with central-type SCLCs, the higher median TMB of peripheral-type SCLCs was observed (P=0.046, Figure 1D). This result indicated that peripheral-type SCLCs might be predictive of the pathological response to immunotherapy.
Full table
Cancer chromosomal instability (CIN) results in an increased rate of change of chromosome number and structure and generates intra-tumor heterogeneity. We calculated wGII to access the CIN status. Tumor CNA burden is the degree to which a cancer genome is altered as a percentage of genome length and represents genome CNA level (31-33), and another indicator of CIN. As was shown in Table 2, there are higher wGII and CNA burden in smoking patients than non-smokers (P=0.0017, Figure 1E and P<0.001, Figure 1F). Furthermore, these alteration of CIN and CNA burden in smoking patients were more obviously in central-types (P=0.00077). Hence, these findings showed that smoking may cause greater damage to chromosomes.
In parallel, it has recently been reported that ITH and SDI, which may reflect the subclonal structures and intra-tumor heterogeneities of tumors, may influence immune surveillance (26,34). There were no significant correlations between ITH or SDI and clinical features (Table 2). Moreover, K-M curves were applied to analyze the correlations between overall survival (OS) and ITH, TMB, SDI, CNA burden, and wGII, but no significant result was found (data not shown).
Driver recurrent SCNAs in Chinese SCLCs
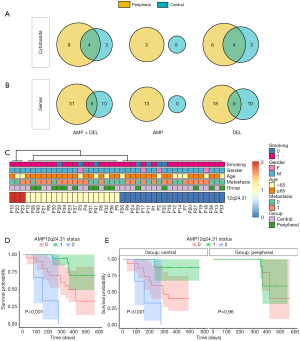
SCNAs play central roles in oncogenesis, cancer therapy and prognosis. It is generally believed that recurrent CNA regions play a role as driver genes (31,32). We applied the GISTIC algorithm, to identify likely driver SCNAs by evaluating the amplitude and frequency of observed events. As shown in Figure 2A,B and Tables 3,4, recurrent CNA regions in central-type and peripheral-type SCLCs had different characteristics. Peripheral and central-type SCLCs had no common recurrent AMP cytobands (peripheral vs. central: 3 vs. 0, Figure 2A middle) or genes (peripheral vs. central: 13 vs. 0, Figure 2B middle). In contrast, the peripheral and central-type SCLCs had common recurrent DEL regions including four cytobands (peripheral vs. common vs. central: 6 vs. 4 vs. 3, Figure 2A right) or five common genes (peripheral vs. common vs. central: 18 vs. 5 vs. 10, Figure 2B right). Interestingly, MYC AMP in 8q24.21 and RB1 DEL 13q14.2, two known driver genes, acted as driver SCNAs of peripheral-type SCLCs (Table 2), indicating the driver SCNAs of central and peripheral-type SCLCs are largely different.

Full table

Full table
We clarified the significance of SCNAs in SCLCs clinical characteristics and prognosis, and analyzed the correlation between OS and central or peripheral-type. The relations between OS and all the CNA regions classified by the GISTIC algorithm were analyzed with K-M curves, and only 12q24.31 AMP was found with a P value <0.05 (data not shown). The status of 12q24.31 and other clinical characteristics are shown in Figure 2C. The AMP CNA of 12q24.31 was significantly related with OS in SCLCs (P=0.00012, Figure 2D), especially in central-type (P=0.0062, Figure 2E), but not peripheral-type. 12q24.31 AMP was not a recurrent CNA in central or peripheral-type SCLCs, and high-level copy number aberrations of 12q24.31 were detected in three central-type samples only, whereas low-level AMPs were in both central and peripheral-type. The correlations between 12q24.31 AMP and all the clinical characteristics were also investigated, but no significant correlation was found (data not shown). Considering these results, it is suggested that SCLCs at different anatomical locations had different biological characteristics.
De novo signatures in Chinese SCLCs
Mutational signatures might reflect DNA damage on tumor genome, which was caused by exposure of DNA-damaging agents. Various carcinogenic and cancer-related processes contribute to the mutational patterns observed in tumors (6,32-35). However, the mutational patterns in Chinese SCLCs remain unknown. We used the R package MutationalPatterns to analyze the de novo signatures of SCLCs, using the NMF algorithm, and found four signatures which were named as Signature A–D (Figure S1A). The 96 base substitutions profiles described the character of Signature A–D (Figure S1B). Using cosine similarity, we assessed the similarities between these four de novo signatures and COSMIC single base substitution signatures (Figure 3A). Signature B is highly similar to COSMIC signatures 4 and 24 (cosine similarity 0.946 and 0.806, respectively), which is believed to be due to smoking status in lung cancer (signature 4) or aflatoxin exposures in liver cancer (signature 24). Signature D is highly similar to COSMIC signatures 6 and 15 (cosine similarity 0.821 and 0.803, respectively), which was previously identified to be associated with defective DNA mismatch repair or small insertions and DELs at mono/polynucleotide repeats. The mutational contributions of each de novo signatures per SCLC patient was examined. We found that the absolute contribution of signature A was significantly higher in peripheral than central-type SCLCs (P=0.03, Table S2). Furthermore, we observed the relationship between clinical features and mutational patterns (Table S2). Signature A was significantly higher in peripheral SCLC patients whose age below 65 (P=0.0091), and male peripheral SCLC patients (P=0.022). However, no significantly differences of the relative contributions were found between the two types of SCLCs when stratified by clinical characteristics or not, except signature B in non-smokers (P=0.037, data not shown).
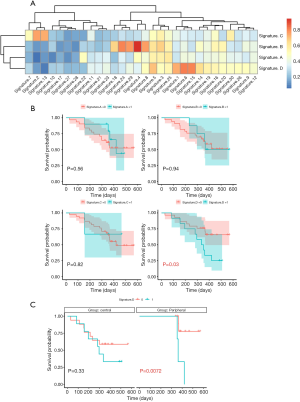
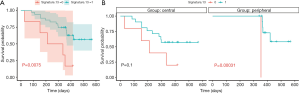
Next, we further analyzed the relationship between mutation profiles and OS. To reduce false positives in signature fitting, a correction was applied to the approach, i.e., a signature’s contribution to a sample was considered as negative if it accounts for <5% of the sample’s mutations, similar to the deconstructSigs package. As was shown in Figure 3B, Signature D was predictive of poor outcome (P=0.03), especially in peripheral SCLC patients (P=0.0072, Figure 3C). In addition, the relationship between all clinical factors and OS was analyzed using K-M survival curve, but no significant difference was found (Figure S2). The absolute contributions of COSMIC signatures between central and peripheral-type SCLCs and clinical stratifications were also analyzed. Signature 3 and 12 were significantly higher in peripheral than central-type SCLCs (P=0.025 and P=0.039, data not shown), especially patients whose age below 65 (P=0.016 and P=0.03), and male (P=0.042 and P=0.073). Signatures 21–23 was significantly higher in peripheral SCLC patients whose age below 65 (P=0.0016, P=0.036, and P=0.0016). Moreover, signature 13 was found with better OS (P=0.0075, Figure 4A), especially in peripheral-type SCLCs (P 0.00031, Figure 4B). No significant correlation was found between OS and the other COSMIC signatures (Figure S3). Altogether, the discoveries of de novo signatures of SCLCs indicated that mutational signatures SCLCs at distinct anatomical locations had different molecular markers and prognostic indicators.
Discussion
Here we provide a comprehensive analysis of somatic genome alterations in SCLC, identifying genomic landscape of Chinese SCLCs and the correlations between SMG, TMB, CIN, CNA burden and clinical characteristics. This is the first study that clarified distinct patterns of somatic genome alterations in in central and peripheral-type SCLC. Although central and peripheral-type SCLCs had similar SMGs, TMB of peripheral-type SCLCs were higher than central-type. As well known, the leading hypothesis in the immunotherapy field is that tumors with high TMB present more neoantigens, and are more immunogenic (26,36). The discovery indicated that peripheral-type SCLCs might be associated with an enhanced response to immune checkpoint blockade therapy. In general, wGII reflected cancer CIN status, and tumors with more CNA burden exhibited mutations in DNA break repair genes (31-33). We found that smoking patients had significantly higher wGII and CNA burden than the non-smokers in SCLCs. Hence, these findings showed that smoking causes greater damage to CIN, and provide a novel insight into the effect of tobacco in SCLC tumorigenesis.
It was well demonstrated that specific SCNAs are associated with clinical characteristics and prognosis of cancers, as well as the potential driver genes they contain (35,37,38). Interestingly, we uncovered that driver SCNAs between central and peripheral-type had their own characteristics. AMP of 8q24.21, which contains the MYC gene, and DEL of 13q14.2, which contains the RB1 gene, act as driver SCNAs in peripheral-type SCLCs. Moreover, the AMP CNA of 12q24.31 is related with OS in central but not peripheral-type SCLCs. These results suggest that driver SCNAs of different anatomical locations lead to distinct prognostic indicators.
Our de novo signatures of SCLCs analysis observed that signature A was significantly higher in peripheral than central-type SCLCs, and signature D was predictive of poor outcome especially in peripheral SCLC. COSMIC signatures attributed to de novo signatures, having similar relationship with SCLCs. These data revealed novel molecular marker and prognostic indicator for SCLCs. Surprisingly, signature 4 contribution was higher in non-smokers, which was identified higher in smokers with lung cancer in previous work. Strong activity of signature 4 was found in many smokers with LADC (6,32,33,35,39). However, about one-third of smoker LADCs displayed minor to no contribution of signature 4 despite their smoking history indicating that smoking-associated mutagenesis minimally contributed to their mutational history (40). Although most SCLC patients were current or former heavy smokers, smoking was associated with increased mutation burdens such as CIN and CNA burden (Figure 1C,D), but not signature 4. It was an interesting to analyze the negatively correlation between signature 4 and SCLCs.
Conclusions
We had provided the first comprehensive genomic analysis of central and peripheral-type SCLCs. Although central and peripheral-type SCLCs had similar SMGs, immunotherapy response, genome instability, the driver SCNAs and mutational signatures were different. These mutations leaded to differences effect in immunotherapy, tumorigenesis and prognosis between central and peripheral-type SCLCs. Our research established novel tumor markers, therapeutic targets and prognostic indicators for SCLCs.
Acknowledgments
Thanks for HaploX Biotechnology, Co.’s technical support of NGS.
Funding: Supported by National Natural Science Foundation of China (81871865), Science and technology innovation action plan of Shanghai Science and Technology Commission (19411950300 and 19411950301), Collaborative innovation plan of Shanghai Health Committee (2020CXJQ02), Key Disciplines of Oncology in Shanghai Pulmonary Hospital.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-1096
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-1096
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-1096). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experimental plans and protocols for this study (No. K18-203Y) were approved by the ethics/licensing committee of the Shanghai Pulmonary Hospital for review and confirmation. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from all patients participating in the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rudin CM, Poirier JT. Small-cell lung cancer in 2016: shining light on novel targets and therapies. Nat Rev Clin Oncol 2017;14:75-6. [Crossref] [PubMed]
- Gazdar AF, Bunn PA, Minna JD. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat Rev Cancer 2017;17:725-37. Erratum in: Nat Rev Cancer 2017;17:765. [Crossref] [PubMed]
- Kanaji N, Sakai K, Ueda Y, et al. Peripheral-type small cell lung cancer is associated with better survival and higher frequency of interstitial lung disease. Lung Cancer 2017;108:126-33. [Crossref] [PubMed]
- George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47-53. [Crossref] [PubMed]
- Govindan R, Ding L, Griffith M, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 2012;150:1121-34. [Crossref] [PubMed]
- Alexandrov LB, Ju YS, Haase K, et al. Mutational signatures associated with tobacco smoking in human cancer. Science 2016;354:618-22. [Crossref] [PubMed]
- Rudin CM, Poirier JT, Byers LA, et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer 2019;19:289-97. [Crossref] [PubMed]
- Pleasance ED, Stephens PJ, O'Meara S, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 2010;463:184-90. [Crossref] [PubMed]
- Waqar SN, Morgensztern D. Treatment advances in small cell lung cancer (SCLC). Pharmacol Ther 2017;180:16-23. [Crossref] [PubMed]
- Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 2012;44:1111-6. [Crossref] [PubMed]
- Peifer M, Fernandez-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104-10. [Crossref] [PubMed]
- Miyauchi E, Saito Y, Ninomiya H, et al. Distinct characteristics of small cell lung cancer (SCLC) correlate with central or peripheral origin - microarray analysis of snap frozen tissue samples. J Thorac Oncol 2013;8:S740.
- Miyauchi E, Motoi N, Ono H, et al. Distinct characteristics of small cell lung cancer correlate with central or peripheral origin: subtyping based on location and expression of transcription factor TTF-1. Medicine (Baltimore) 2015;94:e2324 [Crossref] [PubMed]
- Bandoh S, Fujita J, Ueda Y, et al. Expression of carcinoembryonic antigen in peripheral- or central-located small cell lung cancer: its clinical significance. Jpn J Clin Oncol 2001;31:305-10. [Crossref] [PubMed]
- Slater PM, Grivell R, Cyna AM. Labour management of a woman with carnitine palmitoyl transferase type 2 deficiency. Anaesth Intensive Care 2009;37:305-8. [Crossref] [PubMed]
- Tarasov A, Vilella AJ, Cuppen E, et al. Sambamba: fast processing of NGS alignment formats. Bioinformatics 2015;31:2032-4. [Crossref] [PubMed]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. [Crossref] [PubMed]
- Kim S, Scheffler K, Halpern AL, et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods 2018;15:591-4. [Crossref] [PubMed]
- Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 2016;44:e131 [Crossref] [PubMed]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164 [Crossref] [PubMed]
- Sanchez-Vega F, Mina M, Armenia J, et al. Oncogenic signaling pathways in the Cancer Genome Atlas. Cell 2018;173:321-37.e10. [Crossref] [PubMed]
- Mayakonda A, Lin DC, Assenov Y, et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018;28:1747-56. [Crossref] [PubMed]
- Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214-8. [Crossref] [PubMed]
- Mermel CH, Schumacher SE, Hill B, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011;12:R41. [Crossref] [PubMed]
- Burrell RA, McClelland SE, Endesfelder D, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature 2013;494:492-6. [Crossref] [PubMed]
- Wolf Y, Bartok O, Patkar S, et al. UVB-induced tumor heterogeneity diminishes immune response in melanoma. Cell 2019;179:219-35.e21. [Crossref] [PubMed]
- Roth A, Khattra J, Yap D, et al. PyClone: statistical inference of clonal population structure in cancer. Nat Methods 2014;11:396-8. [Crossref] [PubMed]
- Blokzijl F, Janssen R, van Boxtel R, et al. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med 2018;10:33. [Crossref] [PubMed]
- Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics 2010;11:367. [Crossref] [PubMed]
- Kassambara A. ggpubr: 'ggplot2' Based Publication Ready Plots. 2018. Available online: http://www.sthda.com/english/rpkgs/ggpubr
- Ji W, Wang W, Zhao X, et al. Homologous recombination within the spike glycoprotein of the newly identified coronavirus may boost cross-species transmission from snake to human. J Med Virol 2020. Available online: https://search.bvsalud.org/global-literature-on-novel-coronavirus-2019-ncov/resource/en/covidwho-17
- Chang J, Tan WL, Ling ZQ, et al. Genomic analysis of oesophageal squamous-cell carcinoma identifies alcohol drinking-related mutation signature and genomic alterations. Nat Commun 2017;8:15290. [Crossref] [PubMed]
- Witkiewicz AK, McMillan EA, Balaji U, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun 2015;6:6744. [Crossref] [PubMed]
- Andor N, Graham TA, Jansen M, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med 2016;22:105-13. [Crossref] [PubMed]
- Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016;48:607-16. [Crossref] [PubMed]
- Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202-6. [Crossref] [PubMed]
- Rheinbay E, Nielsen MM, Abascal F, et al. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature 2020;578:102-11. [Crossref] [PubMed]
- ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 2020;578:82-93. [Crossref] [PubMed]
- Alexandrov LB, Kim J, Haradhvala NJ, et al. The repertoire of mutational signatures in human cancer. Nature 2020;578:94-101. [Crossref] [PubMed]
- Lee JJ, Park S, Park H, et al. Tracing oncogene rearrangements in the mutational history of lung adenocarcinoma. Cell 2019;177:1842-57.e21. [Crossref] [PubMed]


