
MicroRNA expression profiling and biomarker validation in treatment-naïve and drug resistant non-small cell lung cancer
Introduction
Lung cancer is the leading cause of cancer-related death worldwide, accounting for 18.4% of cancer deaths in 2018 (1,2). Non-small cell lung cancer (NSCLC) accounts for approximately 85% of lung cancers (3,4). NSCLC consists of two main histological subtypes, adenocarcinoma (ADC) and squamous cell carcinoma (SqCC). Large cell carcinoma (LCC) is largely considered a pathological diagnosis of exclusion and accounts for <10% of cases (5). Despite significant advances in the management of NSCLC over the past number of years as a result of biomarker-driven therapy, 5-year survival rates remain poor at <16%. This is attributable, at least in part, to the emergence of acquired drug resistance during treatment (4). The high mortality rate associated with NSCLC is a result of the absence of suitable screening programmes (6), a lack of emerging personalised therapeutics beyond targeted therapies such as EGFR (7) and ALK inhibitors (8) and the current inability to stratify patients based on inherent or acquired resistance to conventional chemotherapeutic drugs prior to, and during, chemotherapy. Since its FDA-approval, cisplatin has represented a major landmark in the success of anti-cancer therapeutics (9). Although NSCLC patients show an initial response to cisplatin, the majority of patients eventually relapse. The identification of biomarkers that can stratify patients into responders or non-responders, thereby avoiding toxic side-effects associated with platinum-containing regimens, could be of great clinical benefit to lung cancer patients (10).
There exists a significant body of evidence linking the dysregulation of miRNAs with the initiation and progression of lung cancer, in addition to the clinical application of miRNAs in the diagnosis and prognosis of this cancer type (11). More recently, the aberrant expression of these miRNAs has been associated with therapeutic resistance (12). Elucidating the role of miRNAs as tumour suppressors and oncogenes may identify potential therapeutic targets in the treatment of lung cancer. The same is true for identifying a role for miRNAs in therapeutic resistance, the targeting of which, may induce sensitivity to current therapies. To date, a number of studies have reported a link between miRNAs and resistance to chemotherapeutic drugs and radiotherapy in different cancer types, including lung cancer (13). In a study examining the ability of exosomes to increase cisplatin resistance of recipient cells, miRNA expression profiles of A549 cells and exosomes derived thereof, identified the downregulation of miR-100-5p as a key miRNA in the development of resistance via regulation of its target, mammalian target of rapamycin (mTOR). Exosomes were shown to confer resistance of recipient cells to cisplatin via an exosomal miR-100-5p-dependent manner in vitro and in vivo (14). Expression profiling of miRNAs in two drug-resistant NSCLC cell lines [cisplatin resistant (CisR) A549 cells and gefitinib resistant HCC827 cells] identified 29 miRNAs that were upregulated and 36 miRNAs that were downregulated, the majority of which were found to be involved in Ras signalling (15). In NSCLC patients harbouring EGFR mutations, Yin et al. reported the clinical significance of let-7 and miR-17 in resistance to gefitinib in PC9 resistant cells (16). Mechanisms regulating radiation resistance in NSCLC have also been reported and have shown that overexpression of miRNAs such as miR-9, increases radiosensitivity of A549 cells via inhibition of cell proliferation, apoptosis and migration. It was found that the promoter methylation status of miR-9 was also increased in response to ionising radiation (17).
In addition to their functional role in cancer initiation, progression and response to anti-cancer therapeutics, we hypothesise that miRNAs may have diagnostic, prognostic and predictive potential. This study investigated the functional role and biomarker potential of miRNAs in NSCLC using a unique isogenic in vitro model of cisplatin resistance representing all NSCLC histological subtypes, formalin-fixed paraffin-embedded (FFPE) tumour tissues derived from a xenograft model of resistance, in addition to examining the translational relevance of these miRNAs in matched normal and tumour lung tissues and sera from ADC and SqCC patients.
Methods
Cell culture
The LCC H460 parental (PT) and its CisR subline were donated by Dr Dean Fennell, Queen’s University Belfast. The H1299 ADC cell line and its CisR subline were gifted from Dr Parviz Behnam-Motlagh, Umea University, Sweden. The MOR (ADC) cell line and its corresponding CisR subline, in addition to the A549 (ADC) and SKMES-1 (SqCC) cell lines were purchased from the American Type Culture Collection (LGC Promochem, UK). CisR sublines were generated from each PT cell line by continuous cisplatin exposure, as described (18). Cell lines were authenticated using the PowerPlex® 16 HS System (Source BioScience, UK) and maintained in a humidified atmosphere of 5% CO2 at 37 °C (18).
RNA isolation
Total RNA was isolated from cell lines and serum using TRI Reagent (Molecular Research Center, USA) and TRI Reagent Blood, respectively. Following total RNA isolation, small RNAs were isolated using the miRCURY RNA isolation kit (Exiqon, Denmark) and the miRCURY RNA FFPE isolation kit (Exiqon) for FFPE tissues. RNA was quantified using a Nanodrop 1000 spectrophotometer.
MicroRNA (miRNA) profiling
MiRNA profiling was carried out using 7th generation miRCURY LNA miRNA in-situ hybridisation arrays (Exiqon). The samples were labelled using the miRCURY LNA miRNA Hi-Power Labelling Kit Hy3/Hy5 and hybridised on the miRCURY LNA™ miRNA Array. The miRPlus™ sequences are in-licensed human candidate miRNA sequences consisting of in-licensed miRNAs, cloned sequences (Exiqon) and miRNA sequences found in literature searches, many of which are proprietary to Exiqon and are not yet annotated in miRBase. The sequences have been subjected to strict quality control for validity.
Quantitative Real-Time qPCR
RNA (20 ng) was reverse transcribed to cDNA using the Universal cDNA synthesis kit II (Exiqon) and qPCR reactions were completed using the ExiLENT SYBR green master mix and LNA primers. miRNA analyses and validations were carried out by qPCR using the 2−∆∆Ct fold-change relative quantification criteria. As such, the CT threshold was set within the exponential phase of amplification and data were normalised using the appropriate miRNA endogenous control, small nuclear RNA U6. Two miRNAs, miR-103a-2-5p and miR-191-5p, were used as a quality control to determine red blood cell lysis in serum.
Transfections
Transient overexpression of miR-4286 was carried out by transfection of pre-miR-4286 or scrambled control miRNA precursor (Ambion) using FuGENE HD transfection reagent (Promega). MiRNAs belonging to the miR-30 family and miR-34a-5p were inhibited by transient transfection using miRCURY miRNA inhibitors (Exiqon). For inhibition of the miR-30 family, transfection of a combination of three oligonucleotides that could target and inhibit the activity of the miR-30 family (miR-30a-3p, miR-30b-5p and miR-30c-5p) was used. Cells were transfected at a density of 3×105 cells/well for 72 h.
Proliferation
Cell proliferation was measured by BrdU ELISA (Roche Diagnostics Ltd., UK). Cells were treated for 72 h with cisplatin (0–100 µM). Absorbance was recorded at 450 nm and sensitivity to cisplatin was calculated.
Clonogenic survival
Cells were seeded at optimal cell densities and treated with increasing concentrations of cisplatin (0–10 µM) for 72 h, after which time media was replaced with treatment-free media and further incubated for 10 days. Colonies were fixed and stained with 25% (v/v) methanol, 0.05% (w/v) crystal violet. Colonies were counted using the ColCountTM (Oxford Optronix Ltd, UK).
Apoptosis
NSCLC cells (1×105 cells/well) were seeded in 6-well plates and treated for 48 h with increasing concentrations of cisplatin (0–100 µM). Both adherent and floating cells were collected and stained using Annexin V-FITC (IQ Products, Netherlands) and 1 µg/mL propidium iodide (Invitrogen). Flow cytometry analysis was used to quantify apoptotic cells using a Dako CyAn™ADP flow cytometer.
Patient samples
Informed consent was obtained from all patients. Resected tissues and matched sera from NSCLC patients were obtained from the Lung Cancer Biobank at St. James’s Hospital, Dublin. Ten age- and sex-matched normal sera were purchased from BBI Solutions (Cardiff, Wales). Patient demographics and tumour characteristics were recorded (Table S1).
In silico analysis
MiRNAs of interest were analysed in different lung cancer datasets using The Cancer Genome Atlas (TCGA). This consisted of 775 data sets across which miRNA expression (miR-30a-3p, miR-30b-5p, miR-30c-5p, miR-34a-5p, miR-4286) was examined in matched normal and tumour lung tissues from adenocarcinoma (normal, n=44; tumour, n=447) and SqCC (normal, n=44; tumour, n=336) patients. MiRNAs of interest were analysed in different lung cancer datasets (LUAD and LUSC) using KM-Plot at default settings to assess if expression is associated with survival benefit (19). Potential targets for miRNAs were identified using Targetscan (Release 7.2) (20). Of these targets, the top 10 genes associated with cancer cell proliferation were examined by meta-analysis for changes in expression in SqCC using Lung Cancer Explorer (LCE) (21). Gene expression differences between matched normal and tumour tissues were calculated by standardised mean differences (tumour-normal) using Hedges’ G as an effect size metric. For this, all studies included for meta-analysis must have at least 10 samples in each group and meta-analysis was only performed for genes with data available from at least three qualifying studies. Survival association with gene expression are based on univariate Cox Proportional-Hazards modelling. For each study included, gene expression was normalised to zero mean and unit variance for each gene before the model was fitted. All analyses were carried out using the standard default settings.
In vivo tumour formation
Female 7–9 week old NOD/SCID (NOD.CB17-Prkdcscid/NCrCrl) mice were obtained from Charles River Laboratories. Mice were randomised into groups (n=4/group) and subcutaneously injected above the right hind-limb with 1×103 cells (H460 PT or CisR) within a Matrigel plug. Mice were monitored and tumour volumes recorded using digital callipers. Tumour volumes were calculated using the modified ellipsoid formula. Once the experimental endpoint was reached, animals were sacrificed, tumours harvested and preserved in FFPE blocks.
Statistical analysis
Analysis between groups was tested using analysis of variance (ANOVA). Statistical comparison of two means was carried out using an unpaired two-tailed Student’s t-test. Survival analysis between high and low miRNA expressors was performed using the Gehan-Breslow Wilcoxin test. Statistical testing of the in vivo xenograft model was carried out using the non-parametric Mann-Whitney U test. Significance was defined as P≤0.05. Data are represented as Mean ± Standard Error of the Mean (SEM). All data were analysed using GraphPad Prism™ statistical software.
Ethics
Patient samples used in this study were ethically approved by the St James’s Hospital & Tallaght University Hospital Joint Research Ethics Committee (Ref. No. 041018/8804). All animal research work was carried out in the Comparative Medicine Unit, Trinity College Dublin, a Health Products Regulatory Authority (HPRA) approved establishment that operates in accordance with Directive 2010/63/EU and its Irish transposition S.I No. 543 of 2012. Trinity College Dublin complies with the Council for International Organisations of Medical Sciences’ (CIOMS), International Guiding Principles for Biomedical Research Involving Animals, and all laws, regulations and policies governing the care and use of laboratory animals in the jurisdiction in which the research is being conducted. This animal study was approved by the Animal Research Ethics Committee (AREC) at Trinity College Dublin (Ref B100/4400) and carried out under licence B100/4400 from the Health Products Regulatory Authority (HPRA) in Ireland (2011 & 2016).
Results
CisR NSCLC cells exhibit an altered miRNA expression profile
Cisplatin resistance was confirmed in an isogenic panel of parental (PT) and CisR NSCLC cell lines previously established in our laboratory (18). Significantly greater IC50 concentrations were determined for each CisR subline relative to its PT counterpart (P<0.001) (Figure 1A) in response to increasing concentrations of cisplatin following dose-response studies.
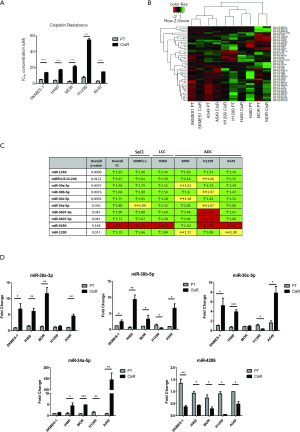
miRNA profiling of PT and CisR cell lines was carried out using 7th generation miRCURY LNA miRNA in-situ hybridisation arrays (Figure 1B). The miRNA profiling used (Exiqon) offers full coverage of miRBase 18 on the 7th generation slides and all probes were validated to hybridise specifically to the sequences in miRBase. However, it has been found that a small number of probes display atypical signal patterns such as very high signal levels in many different sample types. As such, these signals may not represent biologically relevant miRNA expression. Therefore, to avoid focusing on potential false-positives, a total of 22 miRNAs were excluded from the unsupervised analysis shown in the heat map. These miRNAs were however included in the supervised analysis. miRNAs that were excluded are as follows: hsa-miR-1246, hsa-miR-1273g-3p, hsa-miR-1280, hsa-miR-1908, hsa-miR-3124-3p, hsa-miR-3686, hsa-miR-3940-5p, hsa-miR-3960, hsa-miR-4279, hsa-miR-4285, hsa-miR-4290, hsa-miR-4443, hsa-miR-4454, hsa-miR-4456, hsa-miR-4467, hsa-miR-4497, hsa-miR-4516, hsa-miR-4639-3p, hsa-miR-4708-3p, hsa-miR-4764-3p, hsa-miR-4787-5p, hsa-miR-4800-3p, hsa-miR-5100. Profiling of 2,086 miRNAs using in situ hybridisation, identified 10 miRNAs that were most significantly altered between PT and CisR cell lines. The criteria used for cut-off for subsequent validation by qPCR was based on (I) an overall fold-change in expression of >1.4 which was considered significant, and (II) fold-changes in expression that were consistent across a minimum of four out of five cell lines (Figure 1C).
Validation of these 10 miRNAs identified from miRNA profiling arrays were validated by quantitative real-time PCR using locked nucleic acid (LNA) detection primers where miRNA expression was defined within each PT and CisR cell line (SKMES-1, H460, MOR, H1299 and A549) with a CT value <35. Based on our selection criteria above, five of these miRNAs failed on the basis of either having a CT value >35 or were undetectable by qPCR. As such, a 5-miR signature was deduced, representing miRNAs significantly altered between sensitive and resistant cell lines and included; miR-30a-3p, miR-30b-5p, miR-30c-5p, miR-34a-5p and miR-4286 (Figure 1D). Expression of the miR-30 family was altered across at least four out of the five NSCLC cell lines examined (SKMES-1, H460, MOR, H1299 and A549) and represent the different NSCLC histologies of SqCC (SKMES-1), LCC (H460) and adenocarcinoma (MOR, H1299, A549). While miR-1246 was shown to be consistently increased across all paired NSCLC cell lines based on miRNA array data (Figure 1C), validation by qPCR showed no detectable expression of miR-1246, miRPLUS-1246, miR-3607-5p or miR-1290 in the panel of cell lines examined. Furthermore, expression of miR-3607-3p and miRPLUS-G1246 were significantly altered in only one (H1299) and two (H1299 and A549) cell line pairs, respectively. As such, based on our inclusion criteria, 5 miRNAs validated as being significantly altered across our panel of PT and CisR cell lines (miR-30a-3p, miR-30b-5p, miR-30c-5p, miR-34a-5p and miR-4286).
As with all types of analyses involving arrays, validation of altered genes is critical in order to account for any artifact’s that may arise that may potentially lead to skewed data. Due to the high number of genes being tested in parallel, microarrays are prone to false-positive results. Upon validation by qPCR, differences in expression of miRNAs such as miR-34a-5p and miR-4286 do reflect data obtained from the micro-profiling array, hence the necessity to validate the array data by miRCURY LNA™ Universal RT miRNA PCR. While it is possible to validate miRNAs that show small changes in expression, it is important to note that smaller fold-changes tend to be relatively more affected by technical variance. Such changes are thus associated with increased risk of false-positive signals. It is also a possibility that poor correlation between array and qPCR results may arise if the RNA samples are degraded. Most miRNAs however, will be appropriately detected and unaffected by degradation by both qPCR and array. In some situations, miRNAs can be partially degraded with as little as 1–2 end nucleotides. Typically, array capture probes will detect such partially degraded miRNAs whereas the miRCURY™ LNA Universal RT miRNA qPCR primers will discriminate miRNAs that are one or more nucleotides shorter. In this instance, qPCR and array results may not correlate as well. Furthermore, for degraded RNA samples, fragments of larger RNA species that contain miRNA-like sequences may start to hybridise to the capture probes. There are several miRNAs that have great sequence similarity to, for example, tRNAs (miR-1280, miR-1275, miR-720). However, the nature of the qPCR assay design will not detect such miRNA-like sequences present on either intact or degraded fragments of larger RNA species and consequently, qPCR and array date will not correlate as well for such miRNAs. Under ideal conditions, one would expect such perfect correlations. By incorporating redundancy into our criteria for validation, this issue was reduced. For miR-4286, our data show that this miRNA is in fact downregulated across all cell lines (Figure 1D) in contrast to the non-perfect array-based correlation (Figure 1C), clearly demonstrating that this miRNA is indeed important and associated with the development of resistance to cisplatin.
Manipulation of miRNA expression does not re-sensitise NSCLC cells
To investigate the functional role of the miR-30 family members and miR-34a-5p in modulating cisplatin resistance, expression of these miRNAs were transiently inhibited in the H460 CisR subline. Conversely, miR-4268 was transiently overexpressed. Following transfection, cisplatin resistance was investigated across a number of functional parameters relative to scrambled controls to determine the potential sensitising effect of this miRNA modulation. Altered expression was confirmed by qPCR.
Transient inhibition of the miR-30 family (Figure 2A) resulted in a significant downregulation of miR-30b-5p and miR-30c-5p. Following this inhibition, cell proliferation was assessed in response to cisplatin. No significant difference in IC50 concentrations were observed between the miR-30 family and corresponding scrambled controls (P=0.1996). While no significant differences were observed in apoptosis, clonogenic survival was significantly decreased at lower concentrations of cisplatin (0.1–1 µM) following transient inhibition of the miR-30 family relative to scrambled controls. Antisense inhibition significantly reduced expression of miR-34a-5p (P<0.0001) (Figure 2B). Downregulation of miR-34a-5p had no effect on the proliferative capacity of the H460 CisR subline when challenged with cisplatin with no significant difference observed between IC50 cisplatin concentrations (P=0.8840). Similarly, transient inhibition of miR-34a-5p induced no tumour cell apoptosis but significantly decreased clonogenic survival (0.1–2.5 µM).
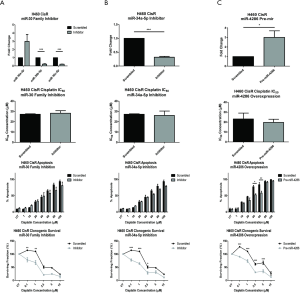
Pre-miR transfection of miR-4286 induced a significant 3.00±0.68-fold upregulation in expression of H460 CisR cells relative to scrambled controls (Figure 2C). No significant difference in IC50 cisplatin concentrations following transfection with pre-miR-4286 was observed. However, miR-4286 overexpression decreased apoptotic cell death when treated with cisplatin (40–60 µM) and decreased clonogenic survival across a range of cisplatin concentrations (0.1–5 µM). Taken together, these data suggest that while modulation of miRNA expression in cisplatin resistant NSCLC may not alter cisplatin sensitivity or enhance cisplatin-induced apoptosis, miRNAs may play a role in the survival capacity of resistant cells.
NSCLC tumours display altered expression of a 5-miR panel
Cisplatin-associated miRNAs identified in our panel of cisplatin-resistant NSCLC cell lines relative to their matched parental (sensitive) counterparts were examined as potential biomarkers to differentiate lung tumours from normal lung tissues, in addition to their use in differentiating between NSCLC tumour histologies in pre-treatment fresh-frozen tissues. To ascertain their biomarker potential, miRNA expression was examined in matched normal and tumour lung tissues from ADC patients (Figure 3A), SqCC patients (Figure 3B) and between each tumour histology (ADC versus SqCC) (Figure 3C).
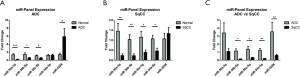
Expression of four of the five miRNAs was significantly altered in ADC tumour tissue relative to normal lung tissue (Figure 3A). Each of the miR-30 family members were significantly downregulated in ADC tumour tissue; miR-30a-3p (0.18-fold, P=0.0007), miR-30b-5p (0.54-fold, P=0.0484) and miR-30c-5p (0.53-fold, P=0.0225). Significant overexpression of miR-4286 was observed in adenocarcinoma tissue relative to adjacent normal lung tissue (5.87-fold, P=0.0477). Reduced expression of the 5-miR signature was also observed in SqCC tumour samples relative to matched normal lung tissues (Figure 3B). Again, a significant downregulation of the miR-30 family of miRNAs was observed in SqCC tumour tissue with a 0.23-fold decrease in miR-30a-3p (P=0.0058), 0.22-fold decrease in miR-30b-5p (P=0.0076) and a 0.38-fold decrease in miR-30c-5p (P=0.03). miR-34a-5p was shown to be significantly downregulated by 0.46-fold (P=0.0136) in SqCC tumours relative to normal lung tissue.
To assess the potential of this miRNA signature in distinguishing between ADC and SqCC histologies, expression within SqCC tissue was examined relative to ADC tissue. There was significantly decreased expression of miR-30b-5p (P=0.0143), miR-30c-5p (P=0.0173), miR-34a-5p (P=0.0018) and miR-4268 (P=0.0250) in SqCC tissues when compared to ADC tissues. Cumulatively, these data suggest that this 5-miR panel has the potential to distinguish ADC and SqCC pathology from that of normal lung tissues, and in discriminating between ADC and SqCC. When miRNA expression in ADC and SqCC tissues and normal lung tissues was compared across lung cancer data sets using The Cancer Genome Atlas (TCGA), similar trends in expression were found in lung tumour tissues for most miRNAs of interest, particularly in SqCC tissues (Table S2). Notably, in both ADC and SqCC tissues, miR-30a-3p and miR-30c-5p were significantly downregulated in both data sets, in contrast to miR-4286 which remained unchanged in both tumour subtypes in the TCGA data set.
While we appreciate that normal and tumour tissues examined as part of this study were not micro-dissected, we are unable to clarify whether normal tissue was present in the tumour samples. This is a common issue with many of such studies and is therefore one limitation of this analysis. Procurement of normal lung tissue following resection was carried out in such a way that tissue was collected at a significant distance from the tumour site. Validation using a larger cohort of patient specimens with appropriate micro-dissection/determination of cancer cell percentages from multiple biobanking sites is warranted to further address this.
Decreased miRNA expression is associated with poor survival in SqCC
The aim of this particular analysis was to examine the prognostic potential of our 5 miRNAs in tumour tissue from NSCLC patients. For both ADC and SqCC histologies, patients were stratified into either high or low expressors based on the median value as a cut-off/threshold and correlated with overall survival. While no significant differences were observed between high and low miRNA expression and overall survival in patients with ADC (data not shown), there was a significant association with high expression of miR-30b-5p, miR-30c-5p and miR-34a-5p and poor overall survival in SqCC patients (P=0.0429) (Figure 4). Due to the high correlation of these three miRNAs, one must consider whether the use of each miRNA as independent markers is feasible in this context. While we are unsure if these miRNAs would have redundancy in relation to predictive testing for cisplatin resistance, one must consider all three miRNAs for any future analysis to determine which has the better predictive value for identifying patients likely to develop resistance to platinum-based therapies, or whether improved specificity and sensitivity could be achieved using all three miRNAs.
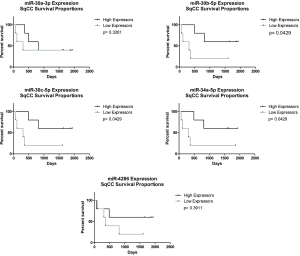
Supporting these findings are miRNA expression and survival data from in silico analysis of lung ADC (n=513) and SqCC (n=478) patients based on miRNA biomarker assessment (Figure S1). These data were collated and statistically analysed by Cox-Regression using online software via the validated pan-Cancer miRNA Kaplan-Meier Plotter. These data showed an association between low expression of members of the 5-miR panel (miR-30b-5p, miR-30c-5p) and poor survival in SqCC. Taken together, these data highlight the potential use of miRNAs, in particular the miR-30 family members, as prognostic biomarkers in lung cancer.
miR-4286 is overexpressed in the sera of SqCC patients
To assess the potential of one or more of these differentially altered miRNAs as a circulating diagnostic biomarker, expression of each miRNA was investigated in the sera of ADC and SqCC patients relative to normal healthy controls (Figure 5A). No detection of miR-30a-3p, miR-30b-5p, miR-30c-5p or miR-34a-5p was observed in sera. A significant 4.63-fold overexpression of miR-4286 was however observed in SqCC serum relative to healthy controls and ADC patients (P=0.0095).
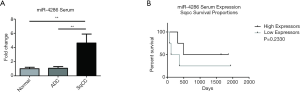
Survival analysis of miR-4286 expression in SqCC sera showed no significant difference (P=0.2330) in survival between high and low expressors (Figure 5B). These data are inconclusive due to the small sample numbers included in this analysis and warrants further validation in a larger cohort of patients. To further assess the potential role of miRNA-4286 in SqCC, TargetScan was used to identify genes which have predicted targets to this miRNA. From this analysis, 4,674 transcripts were identified containing a total of 6,745 sites. To narrow this down, the top 10 mRNA transcripts were selected based on their association with cancer cell proliferation. Systematic meta-analysis demonstrated that 7/10 of the target mRNAs were significantly downregulated in SqCC (Table S3, Figure S2). There was no significant association of these mRNAs with survival in SqCC (Table S4).
Xenograft tumours derived from cisplatin resistant NSCLC cells exhibit increased expression of a 5-miRNA signature
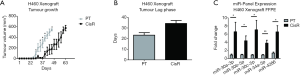
The stable expression of miRNA in FFPE tissues makes these small RNA molecules ideal biological markers in archived specimens. To investigate the expression of the miRNA panel in FFPE preserved lung tumour tissues, an in vivo xenograft model of cisplatin resistant and sensitive tumours was used to assess the expression of the cisplatin-associated miRNAs. H460 PT and CisR sublines were injected into NOD/SCID mice and tumour growth and progression monitored. No significant difference was observed in tumour growth initiation between PT and CisR tumours. Analysis of tumour volume over time (Figure 6A) showed significant differences in tumour growth rate between H460 PT and CisR xenograft tumours. Mean tumour volume of H460 PT xenograft tumours was significantly increased compared to CisR tumours from day 30 (78.38±49.9 vs. 2.75±2.13 mm3, P=0.0286). This is further highlighted by the tumour lag phase between PT and CisR tumours (Figure 6B) during tumour development where it was observed that tumours derived from CisR cells took longer to grow relative to their PT cell counterparts. This pattern of tumour growth mirrors what is observed under cell culture conditions in vitro. Resistant cells grow at a much slower rate and have a greater doubling time compared to parental cells from which they were derived. A significant upregulation of all miRNAs was observed in CisR tumours relative to their PT counterparts (Figure 6C). Combined, these results are indicative of a potential cisplatin-associated 5-miR signature that may have potential use as a biomarker that is predictive of cisplatin response in FFPE tissues following resection of NSCLC tumours in lung cancer patients.
Discussion
Multiple studies have shown a link between miRNAs and cisplatin resistance in NSCLC (14,16,22,23). Mechanisms of chemoresistance and associated molecular targets have been identified in cancer patients using genomic and proteomic analysis. In breast cancer patients, it was reported in a study by Climent et al., that increased sensitivity to anthracycline-based chemotherapy may result from a chromosome 11q deletion containing the sequence for the miRNA, miR-125b (24), suggesting that dysregulation of miRNAs may be linked to drug resistance in cancer. In a NCI panel of 60 human cancer cell lines, there was a strong correlation between miRNA expression and sensitivity to anti-cancer drugs (25) further highlighting a critical role for miRNAs in chemoresistance.
In this study, in-situ hybridisation profiling and qPCR validation identified a 5-miR signature which had the capacity to discriminate between cisplatin sensitive and resistant cells across NSCLC histologies. Within this cisplatin resistant 5-miR signature, four miRNAs, miR-30a-3p, miR-30b-5p, miR-30c-5p and miR-34a-5p, were significantly upregulated while miR-4286 was significantly downregulated across CisR sublines. To date, there have been no studies published demonstrating a link between the upregulation of miR-30 family members (miR-30a-3p, miR-30b-5p and miR-30c-5p) and cisplatin resistance in lung cancer. Previous studies have shown that ectopic expression of miR-30a can enhance the cytotoxic effect of cisplatin in HeLa cells in xenograft models (26). Similarly, forced overexpression of miR-30c increased cisplatin and paclitaxel sensitivity in SKOV-3 and HeyA8-MDR cells, respectively (27).
Inhibition of miR-34a-5p did not sensitise the H460 CisR subline to the anti-proliferative or pro-apoptotic effects of cisplatin; however, the survival capacity was significantly decreased. These results are in agreement with those reported by Wang et al., where inhibition of miR-34a in A549 cells using antisense oligonucleotides did not alter the proliferative or apoptotic response to cisplatin (28). In addition, neither overexpression nor downregulation of miR-34a in SCLC lines influenced sensitivity to cisplatin or etoposide (29). miR-34a has also been found to be associated with cisplatin resistance in the osteogenic sarcoma cell line, Saos-2, where induced overexpression of miR-34a promoted cell survival when challenged with cisplatin (30). Previously, miR-34a has been described as a p53-regulated tumour suppressor miRNA, the levels of which are often decreased in tumour tissue relative to normal lung tissue, similar to what we observed in SqCC tumour tissue. Xue et al. suggested the potential of a miR-34a-based miRNA combination therapy for lung cancer where therapeutic delivery of miR-34a could restore miR-34a expression in lung tumours and enhance its tumour suppressor capacity. Indeed, nanoparticle delivery of miR-34a in combination with cisplatin in mouse models of lung adenocarcinoma significantly slowed tumour growth and decreased tumour volume relative to cisplatin alone (31). Of interest, a number of the target genes regulated by miR-34a include those involved in stemness, such as POU2F1 and Notch, as well as targets that play a key role in the epithelial-mesenchymal transition process (Snail and Notch), a characteristic phenotype associated with resistance (32,33).
Our data demonstrate for the first time that ectopic expression of miR-4286 in cisplatin resistant NSCLC cells (H460) significantly decreased the clonogenic survival of chemoresistant cells. While this modulation of miR-4286 expression did not alter the proliferative capacity of cisplatin-resistant lung cancer cells, its overexpression induced an anti-apoptotic effect in response to treatment with cisplatin. These findings highlight a novel role for miR-4286 in cisplatin resistant lung cancer. Increased expression of miR-4286 has previously been reported in oesophageal adenocarcinoma, providing evidence that tissue miRNA profiles can discriminate oesophageal carcinoma from its metaplastic precursor, Barrett’s oesophagus (34). In melanoma cells where miR-4286 mediates proliferation and apoptosis, the use of a miR-4286 inhibitor altered the mRNA expression of several gene targets implicated in the regulation of cell proliferation and apoptosis (35).
miR-30 is frequently overexpressed in human cancers. In a genetic screen of a lentiviral expression library containing 524 miRNAs, miR-30 was found to abrogate oncogenic-induced senescence through its tumour-suppressing mechanisms involving DNA damage responses (DDR), p53 activation and induction of p16INK4A (36). Using cell lines and mouse models, miR-30 could disrupt senescence and promote cancer by targeting CHD7 and TNRC6A. While CHD7 is a transcriptional coactivator of p16INK4A, TNRC6A is a key component in ras-induced DDR RNA expression and functionality. Increased expression of five miR-30 family members (miR-30a, miR-30b, miR-30c, miR-30d, miR-30e) abrogated the induction of proliferative arrest in BJ fibroblast cells, an effect that was replicated in primary WI-38 human fibroblasts and human mammary epithelial cells. Furthermore, it was found that miR-30c could promote cancer development in a pancreatic model by disrupting senescence induction by oncogenic Kras. Consistent with its role as a tumour suppressor, miR-30 is suppressed by oncogenic signals such as epidermal growth factor (EGF) and hepatocyte growth factor (HGF) and is expressed at low levels in prostate cancer tissues. While a number of epithelial-to-mesenchymal transition (EMT)-associated genes are predicted targets of miR-30, the Ets-related gene (ERG) is the most frequently overexpressed oncogene in prostate cancer (37). This is activated by genomic fusion events between promoter upstream sequences of the androgen regulated gene, TMPRSS2, and coding sequences of ERG. Overexpression of miR-30 in VCaP prostate cancer cells suppresses the EMT phenotype and inhibits cell migration, invasion and cell growth in vitro and in vivo where ERG was shown to be a direct target of miR-30. In colorectal cancer, miR-30-5p has been shown to target USP22 and attenuates the Wnt/β-catenin pathway, negatively regulating stemness and chemoresistance in this cancer type (38). Studies by Ma et al. demonstrated that using genome-wide transcriptional profiling of miRNAs in lung cancer stem cells at different stages, alter expression levels of several tumour-related miRNAs during spheroid formation of SPC-A1 and NCI-H1650 NSCLC cell lines. Furthermore, by targeting the TM4SF1 oncogene, it was demonstrated that miR-30a/c inhibits self-renewal and the tumorigenesis of lung tumour initiating cells, and that low expression of miR-30a/c and high expression of TM4SF1 were independent prognostic factors in NSCLC patients. These data provide new evidence for TM4SF1 as a direct target of miR-30a/c (39). More recently, a link between specific miRNAs and patient prognosis and relapse was reported in lung cancer patients using a miRNA signature identified in tumour tissues and sera that was shown to be predictive of overall survival (40,41). Furthermore, a miRNA panel with diagnostic potential in lung adenocarcinoma was reported in sputum samples (42). Studies have also reported the potential use of miRNAs in distinguishing between the lung cancer subtypes, NSCLC and SCLC (43,44). The ability to assess miRNAs in blood as a non-invasive procedure and their high stability in blood components such as serum and plasma, highlights their potential to be used for screening in the prognostic and diagnostic setting (45).
We have shown in this study that miR-30a-3p is significantly downregulated in both ADC and SqCC tissues relative to normal lung tissues. Similar observations were reported by Kumarswamy et al. where miR-30a was shown to be significantly downregulated in NSCLC tumour tissue relative to adjacent normal lung tissue. Multiple meta-analyses have also reported similar findings regarding miR-30a expression in NSCLC tumour tissue (46-48). Furthermore, miR-30b-5p was shown to be significantly downregulated and indicative of cancerous tissue in both histological subtypes when compared to patient-matched normal tissue. These data support the current literature in which miR-30b was shown to be greater than 2-fold decreased in SqCC tissue relative to normal lung tissue (49). These are in agreement with similar findings for miR-30c-5p (50) and miR-34a-5p (51,52). While miR-4286 was significantly downregulated in adenocarcinoma tissue, its expression has not been previously reported in the context of NSCLC tissue. While the cohort of patient samples used in this study was small, it was large enough to correctly identify significantly altered miRNAs in larger in silico TCGA datasets. However, it must be noted that differences in cell miRNA expression in vitro and in patient lung tumour tissues, reflect the clinical setting from which these are derived. The miR-30 family of miRNAs (miR-30a-3p, miR-30b-5p, miR-30c-5p) in addition to miR-34a-5p were for the most part, significantly upregulated in our isogenic panel of cisplatin resistant NSCLC cell lines. This was in contrast to that observed in ADC and SqCC tissues from the patient cohort used in the current study, in addition to those from TCGA datasets, where these were significantly decreased. Tissue samples were derived from treatment-naïve NSCLC patients while in vitro data are representative of the cisplatin resistant phenotype. In the clinic setting, procuring tumour biopsies from NSCLC patients that have progressed on cisplatin-doublet chemotherapy is difficult. In order to further translate these data into a clinically relevant setting, studies are warranted where NSCLC patients receiving cisplatin-doublet chemotherapy are monitored using serial blood sampling during treatment for changes in expression of these miRNAs. This strategy could potentially be used to validate the potential use of these miRNAs in predicting response to cisplatin-based chemotherapy and the development of resistance to this drug.
When miRNAs were examined between ADC and SqCC tissues, a significantly differentiated expression profile was identified, where a panel of four significantly downregulated miRNAs were observed (miR-30b-5p, miR-30c-5p, miR-34a-5p and miR-4286). While miRNA signatures have previously been documented in the literature that can distinguish between NSCLC subtypes, members of the miR-30 family and a four miRNA panel similar to that reported in the current study, has not previously been reported in this context. Differential expression of miR-34a was previously identified as a biomarker of histological discrimination within a signature reported by Huang et al., where it was also shown to be significantly decreased in SqCC tissue compared to that of ADC (53). Depsite these interesting observations of differential expression of miRNAs between different NSCLC histologies, current histopathological methodologies remain at the forefront in lung tumour tissue diagnoses.
Contrary to previous studies, no serum expression of miR-30a-3p, miR-30b-5p, miR-30c-5p or miR-34a-5p was found in NSCLC patients and healthy controls in this study (54,55). Of interest however, increased expression of miR-4286 was observed in the sera of SqCC patients only, relative to those from normal controls and ADC patients. Based on these findings, we report for the first time, the potential use of miR-4286 as a blood-based NSCLC diagnostic biomarker. Given the elevated expression of this miRNA in SqCC, in silico analyses was used to interrogate if any of its candidate targets were potentially altered in SqCC. From the initial screen to identify candidate genes, the top 10 genes which had known associations with cellular proliferation were selected based on their expression in SqCC. A significant number of these genes were found to be significantly downregulated in SqCC. No associations with this downregulation or survival were observed. Whilst suggestive that this miRNA may be regulating the expression of these genes in SqCC, these results remain to be functionally validated in future studies.
Despite the many successes in determining general mechanisms of miRNA generation and function, localising miRNAs to a given cell type or entity is challenging. It has been reported however that while many miRNAs are ubiquitously expressed, some do show expression specificity or marked differences in concentration between different types of cells (56). In this study, miR-4286 was the only miRNA of our panel that could be detected in the sera of NSCLC patients, most significantly in the sera of SqCC patients. While it is somewhat difficult to deduce the exact source of this miRNA, it does raise the question as to whether its abundance and detection in the blood sera may be a direct result of its source of secretion compared to the other miRNAs examined. The cellular composition of a tissue changes in most disease states. In cancer, malignant cells replace the native epithelium while in inflammatory diseases, the number of infiltrating leukocytes can increase markedly. Exosomes may also play a part in this process. These are small microvesicles that range in size from 30–100 nm (57) and are secreted by a variety of cell types including epithelial cells, T- and B-lymphocytes, mast cells, dendritic cells and neurons, where they carry and transfer proteins and nucleic acids between cells. While miRNA signatures have shown promise as diagnostic biomarkers in unfractionated whole serum, urine, saliva and cerebrospinal fluid, there is still little or no consensus about the relative contribution of exosomal miRNAs to whole serum miRNAs (58). Much of the data in this field have demonstrated that the exosome fraction of serum and saliva is highly enriched in miRNAs, and that the majority of this class of RNA is not freely circulating in human biologic fluids but is enclosed within these microvesicles. It has also been widely documented that in addition to being actively secreted via these extracellular vesicles, miRNAs can also be released by passive leakage from lytic cells and via high-density lipoproteins (59,60). Based on these previous studies, examining exosomal miRNAs may be a more appropriate first step for early biomarker studies to reduce the probability of false-negative results due to the presence of low abundance miRNAs that may not be detected using unfractionated serum or saliva.
In this study, we demonstrate that miR-4286 can be detected in patient sera and moving forward, may have potential as a blood-based marker. One limitation of these findings is the small sample size. In order to further validate miR-4286 as a prognostic and predictive marker, larger studies are required. Sample size is a major consideration in the robust identification of candidate biomarkers. In a previous report by Hernández et al., it was suggested that biomarker discovery experiments are undertaken on the maximum number of samples possible, stating that at least 50 samples are preferred as a starting point in this process. In doing so, this would (I) minimise the effects of over-fitting and (II) improve the quality of performance metrics, thereby saving on significant time and effort that may otherwise be wasted in validating a non-predictive biomarker panel using unique and highly valued samples from specific patient cohorts (61).
While it was not possible to validate these data in NSCLC patients who have progressed on platinum-based therapy due to the development of resistance to cisplatin, we further expanded our investigation to address whether this panel of miRNAs had prognostic value in NSCLC and could potentially translate to a more predictive role in the context of response to cisplatin therapy using sera from treatment-naïve patients. To investigate the expression of these five miRNAs in an in vivo xenograft model of cisplatin resistance, RNA was derived from H460 PT and CisR tumour FFPE sections and miRNA expression examined by qPCR. A 5-miR signature showed a differential expression profile in xenograft tumours derived from H460 PT and CisR cells and was capable of distinguishing between cisplatin sensitive and cisplatin resistant lung tumours. However, in contrast to what was found in cisplatin-resistant NSCLC cell lines in vitro, miR-4286 was significantly increased in xenograft FFPE tumour tissue derived from H460 CisR tumours. One possible explanation for these differences in expression may be related to, at least in part, to the influence of the tumour microenvironment in the mouse xenograft study such as stromal and immune cell infiltrates. As such, miR-4286 may not be a reliable marker in the context of cisplatin resistance. Further studies will be required to determine if serum levels of this miRNA have any predictive or prognostic potential, while determining expression of miR-4286 in the context of FFPE lung tumour samples. The in vitro transfection/modulation of miRNAs and in vivo xenograft models used in this study incorporated the use of the H460 PT and CisR cell lines. This matched cell line pair was selected for a number of reasons; (I) transfection efficiency in this cell line was significantly higher than that observed with other NSCLC cell lines, (II) the doubling times for H460 PT and CisR cells was higher in vitro thereby permitting more experimental replicates to be carried out over a shorter period of time, (III) in previous xenograft models established in our laboratory (62), H460 cells had better uptake in vivo, in which case, cell numbers for injection were previously optimised for use in a similar xenograft model.
Conclusions
In conclusion, our data indicate a novel 5-miR signature in NSCLC. Modulation of these miRNAs did not restore the anti-proliferative or pro-apoptotic effects of cisplatin in a model of chemoresistance. However, these miRNAs do show promise as potential diagnostic biomarkers in NSCLC tissues, where their differential expression could differentiate malignant tissue from normal lung tissue and between ADC and SqCC histologies. Furthermore, our data highlight for the first time, the prognostic significance of these miRNAs and their association with poor overall survival in SqCC patients. While miR-4286 was the only one of five miRNAs detectable in the sera from NSCLC patients, further studies are warranted to validate its use potential as a diagnostic biomarker in NSCLC.
Acknowledgments
The authors would like to acknowledge Andrea Ferguson and Helen Buckley (Clinical Nurse Managers), Cancer Clinical Trials Unit at St. James’s Hospital for their support in consenting patients.
Funding: This work was supported by Molecular Medicine Ireland (MMI) as part of the Clinical & Translational Research Scholars Programme (CTRSP) under Cycle 5 of the Irish Governments Programme for Research in Third Level Institutions (PRTLI) and co-funded under the European Regional Development Fund (ERDF).
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-959
Peer Review File: Available at http://dx.doi.org/10.21037/tlcr-20-959
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-959). Professor Kenneth J. O’Byrne serves as an unpaid editorial board member of Translational Lung Cancer Research. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the St James’s Hospital & Tallaght University Hospital Joint Research Ethics Committee (No.: 041018/8804). Animal experiments were performed under a project license (No.: B100/4400) granted by the Health Products Regulatory Authority (HPRA) in Ireland (2011 & 2016), in accordance with Directive 2010/63/EU and its Irish transposition (S.I No.: 543) of 2012, for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians 2018;68:394-424. [Crossref] [PubMed]
- Ferlay J, Colombet M, Soerjomataram I, et al. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. European Journal of Cancer 2018;103:356-87. [Crossref] [PubMed]
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA: A Cancer Journal for Clinicians 2011;61:69-90. [Crossref] [PubMed]
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Rekhtman N, Travis WD. Large No More: The Journey of Pulmonary Large Cell Carcinoma from Common to Rare Entity. J Thorac Oncol 2019;14:1125-7. [Crossref] [PubMed]
- Goldstraw P, Ball D, Jett JR, et al. Non-small-cell lung cancer. Lancet 2011;378:1727-40. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus Chemotherapy in Advanced ALK-Positive Lung Cancer. N Engl J Med 2013;368:2385-94. [Crossref] [PubMed]
- Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer 2007;7:573-84. [Crossref] [PubMed]
- Giaccone G. Clinical perspectives on platinum resistance. Drugs 2000;59:9-17; discussion 37-8. [Crossref] [PubMed]
- Wu KL, Tsai YM, Lien CT, et al. The Roles of MicroRNA in Lung Cancer. Int J Mol Sci 2019;20:1611. [Crossref] [PubMed]
- Dalmay T, Edwards DR. MicroRNAs and the hallmarks of cancer. Oncogene 2006;25:6170-5. [Crossref] [PubMed]
- MacDonagh L, Gray SG, Finn SP, et al. The emerging role of microRNAs in resistance to lung cancer treatments. Cancer Treat Rev 2015;41:160-9. [Crossref] [PubMed]
- Qin X, Yu S, Zhou L, et al. Cisplatin-resistant lung cancer cell-derived exosomes increase cisplatin resistance of recipient cells in exosomal miR-100-5p-dependent manner. Int J Nanomedicine 2017;12:3721-33. [Crossref] [PubMed]
- Hu S, Yuan Y, Song Z, et al. Expression Profiles of microRNAs in Drug-Resistant Non-Small Cell Lung Cancer Cell Lines Using microRNA Sequencing. Cell Physiol Biochem 2018;51:2509-22. [Crossref] [PubMed]
- Yin J, Hu W, Pan L, et al. let-7 and miR-17 promote self-renewal and drive gefitinib resistance in non-small cell lung cancer. Oncol Rep 2019;42:495-508. [Crossref] [PubMed]
- Wei W, Dong Z, Gao H, et al. MicroRNA-9 enhanced radiosensitivity and its mechanism of DNA methylation in non-small cell lung cancer. Gene 2019;710:178-85. [Crossref] [PubMed]
- Barr MP, Gray SG, Hoffmann AC, et al. Generation and characterisation of Cisplatin-resistant non-small cell lung cancer cell lines displaying a stem-like signature. PloS one 2013;8:e54193 [Crossref] [PubMed]
- Nagy Á, Lánczky A, Menyhárt O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 2018;8:9227. [Crossref] [PubMed]
- Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015;4:e05005 [Crossref] [PubMed]
- Cai L, Lin S, Girard L, et al. LCE: an open web portal to explore gene expression and clinical associations in lung cancer. Oncogene 2019;38:2551-64. [Crossref] [PubMed]
- Zhu W, Xu H, Zhu D, et al. miR-200bc/429 cluster modulates multidrug resistance of human cancer cell lines by targeting BCL2 and XIAP. Cancer Chemother Pharmacol 2012;69:723-31. [Crossref] [PubMed]
- Galluzzi L, Morselli E, Vitale I, et al. miR-181a and miR-630 regulate cisplatin-induced cancer cell death. Cancer Res 2010;70:1793-803. [Crossref] [PubMed]
- Climent J, Dimitrow P, Fridlyand J, et al. Deletion of chromosome 11q predicts response to anthracycline-based chemotherapy in early breast cancer. Cancer Res 2007;67:818-26. [Crossref] [PubMed]
- Blower PE, Verducci JS, Lin S, et al. MicroRNA expression profiles for the NCI-60 cancer cell panel. Mol Cancer Ther 2007;6:1483-91. [Crossref] [PubMed]
- Zou Z, Wu L, Ding H, et al. MicroRNA-30a sensitizes tumor cells to cis-platinum via suppressing beclin 1-mediated autophagy. J Biol Chem 2012;287:4148-56. [Crossref] [PubMed]
- Jia W, Eneh JO, Ratnaparkhe S, et al. MicroRNA-30c-2* expressed in ovarian cancer cells suppresses growth factor-induced cellular proliferation and downregulates the oncogene BCL9. Mol Cancer Res 2011;9:1732-45. [Crossref] [PubMed]
- Wang PY, Li YJ, Zhang S, et al. Regulating A549 cells growth by ASO inhibiting miRNA expression. Mol Cell Biochem 2010;339:163-71. [Crossref] [PubMed]
- Lee JH, Voortman J, Dingemans AM, et al. MicroRNA expression and clinical outcome of small cell lung cancer. PloS One 2011;6:e21300 [Crossref] [PubMed]
- Tan J, Fan L, Mao JJ, et al. Restoration of miR-34a in p53 deficient cells unexpectedly promotes the cell survival by increasing NFkappaB activity. J Cell Biochem 2012;113:2903-8. [Crossref] [PubMed]
- Xue W, Dahlman JE, Tammela T, et al. Small RNA combination therapy for lung cancer. Proc Natl Acad Sci U S A 2014;111:E3553-61. [Crossref] [PubMed]
- Imani S, Wei C, Cheng J, et al. MicroRNA-34a targets epithelial to mesenchymal transition-inducing transcription factors (EMT-TFs) and inhibits breast cancer cell migration and invasion. Oncotarget 2017;8:21362-79. [Crossref] [PubMed]
- MacDonagh L, Gallagher MF, Ffrench B, et al. Targeting the cancer stem cell marker, aldehyde dehydrogenase 1, to circumvent cisplatin resistance in NSCLC. Oncotarget 2017;8:72544-63. [Crossref] [PubMed]
- Drahos J, Schwameis K, Orzolek LD, et al. MicroRNA Profiles of Barrett's Esophagus and Esophageal Adenocarcinoma: Differences in Glandular Non-native Epithelium. Cancer Epidemiol Biomarkers Prev 2016;25:429-37. [Crossref] [PubMed]
- Komina A, Palkina N, Aksenenko M, et al. Antiproliferative and Pro-Apoptotic Effects of MiR-4286 Inhibition in Melanoma Cells. PloS One 2016;11:e0168229 [Crossref] [PubMed]
- Su W, Hong L, Xu X, et al. miR-30 disrupts senescence and promotes cancer by targeting both p16(INK4A) and DNA damage pathways. Oncogene 2018;37:5618-32. [Crossref] [PubMed]
- Kao CJ, Martiniez A, Shi XB, et al. miR-30 as a tumor suppressor connects EGF/Src signal to ERG and EMT. Oncogene 2014;33:2495-503. [Crossref] [PubMed]
- Jiang S, Miao D, Wang M, et al. MiR-30-5p suppresses cell chemoresistance and stemness in colorectal cancer through USP22/Wnt/beta-catenin signaling axis. J Cell Mol Med 2019;23:630-40. [Crossref] [PubMed]
- Ma YS, Yu F, Zhong XM, et al. miR-30 Family Reduction Maintains Self-Renewal and Promotes Tumorigenesis in NSCLC-Initiating Cells by Targeting Oncogene TM4SF1. Mol Ther 2018;26:2751-65. [Crossref] [PubMed]
- Hu Z, Chen X, Zhao Y, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol 2010;28:1721-6. [Crossref] [PubMed]
- Yu SL, Chen HY, Chang GC, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell 2008;13:48-57. [Crossref] [PubMed]
- Yu L, Todd NW, Xing L, et al. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer 2010;127:2870-8. [Crossref] [PubMed]
- Landi MT, Zhao Y, Rotunno M, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res 2010;16:430-41. [Crossref] [PubMed]
- Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006;9:189-98. [Crossref] [PubMed]
- Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer science 2010;101:2087-92. [Crossref] [PubMed]
- Võsa U, Vooder T, Kolde R, et al. Meta-analysis of microRNA expression in lung cancer. Int J Cancer 2013;132:2884-93. [Crossref] [PubMed]
- Guan P, Yin Z, Li X, et al. Meta-analysis of human lung cancer microRNA expression profiling studies comparing cancer tissues with normal tissues. J Exp Clin Cancer Res 2012;31:54. [Crossref] [PubMed]
- Kumarswamy R, Mudduluru G, Ceppi P, et al. MicroRNA-30a inhibits epithelial-to-mesenchymal transition by targeting Snai1 and is downregulated in non-small cell lung cancer. Int J Cancer 2012;130:2044-53. [Crossref] [PubMed]
- Gao W, Lu X, Liu L, et al. MiRNA-21: a biomarker predictive for platinum-based adjuvant chemotherapy response in patients with non-small cell lung cancer. Cancer Biol Ther 2012;13:330-40. [Crossref] [PubMed]
- Xia Y, Chen Q, Zhong Z, et al. Down-regulation of miR-30c promotes the invasion of non-small cell lung cancer by targeting MTA1. Cell Physiol Biochem 2013;32:476-85. Erratum in: Cell Physiol Biochem 2013;32:1124. [Crossref] [PubMed]
- Gallardo E, Navarro A, Vinolas N, et al. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis 2009;30:1903-9. [Crossref] [PubMed]
- Wiggins JF, Ruffino L, Kelnar K, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res 2010;70:5923-30. [Crossref] [PubMed]
- Huang W, Hu J, Yang DW, et al. Two microRNA panels to discriminate three subtypes of lung carcinoma in bronchial brushing specimens. Am J Respir Crit Care Med 2012;186:1160-7. [Crossref] [PubMed]
- Montani F, Marzi MJ, Dezi F, et al. miR-Test: a blood test for lung cancer early detection. J Natl Cancer Inst 2015;107:djv063 [PubMed]
- Cui EH, Li HJ, Hua F, et al. Serum microRNA 125b as a diagnostic or prognostic biomarker for advanced NSCLC patients receiving cisplatin-based chemotherapy. Acta pharmacologica Sinica 2013;34:309-13. [Crossref] [PubMed]
- Haider BA, Baras AS, McCall MN, et al. A critical evaluation of microRNA biomarkers in non-neoplastic disease. PLoS One 2014;9:e89565 [Crossref] [PubMed]
- Heijnen HF, Schiel AE, Fijnheer R, et al. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood 1999;94:3791-9. [Crossref] [PubMed]
- Gallo A, Tandon M, Alevizos I, et al. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One 2012;7:e30679 [Crossref] [PubMed]
- Kosaka N, Iguchi H, Yoshioka Y, et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem 2010;285:17442-52. [Crossref] [PubMed]
- Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654-9. [Crossref] [PubMed]
- Hernández B, Parnell A, Pennington SR. Why have so few proteomic biomarkers "survived" validation? (Sample size and independent validation considerations). Proteomics 2014;14:1587-92. [Crossref] [PubMed]
- Barr MP, Gray SG, Gately K, et al. Vascular endothelial growth factor is an autocrine growth factor, signaling through neuropilin-1 in non-small cell lung cancer. Mol Cancer 2015;14:45. [Crossref] [PubMed]


