
One carbon metabolism in human lung cancer
Introduction
With more than 1.4 million deaths annually, lung cancer is the leading cause of cancer related deaths worldwide (1). In addition to the histological differentiation into small cell lung cancer (SCLC) and non-small cell lung carcinomas (NSCLC) with further differentiation into adenocarcinoma (AC) and squamous cell carcinoma (SQCLC), several genetic subgroups of lung cancer have been discovered. AC is the most common subtype accounting for about 30% of cases and 420,000 deaths per year worldwide. They are characterized by a high number of genomic alterations including 32% activating mutations in the KRAS gene as the largest genetic subgroup, followed by activating mutations in the EGFR gene (11%). Genomic fusions of the ALK, Ros and Ret genes occur in 1–3% of cases. In about 25% of cases, no genetic driver mutations have yet been identified (2). Discovery of activating genetic mutations in the EGFR and ALK kinases has led to a marked improvement in the prognosis of these specific subgroups of patients through the use of targeted kinase inhibitors (3,4). Unfortunately, no similar therapeutic successes have yet been achieved for the other subgroups and most patients still receive conventional chemotherapy (5). The chemotherapeutic drug pemetrexed in combination with platinum based chemotherapies (e.g., cisplatin) is one of the current first-line therapeutic strategies for non-operable pulmonary adenocarcinoma. Pemetrexed, like methotrexate and 5-FU, affects the folate metabolism of tumor cells and leads to clinical response in ~30% of patients with AC (6).
One carbon metabolism (1CM) includes the methionine and folate cycles and plays an important role in nucleotide metabolism and genomic maintenance (7-9). The 1CM metabolic network includes mitochondria and cytoplasm. Five major steps are processed by the key enzymes: D-3-phosphoglycerate dehydrogenase 3 (PGDH3), Methylenetetrahydrofolate Dehydrogenase 1 and 2 (MTHFD2/1), Serine methyltransferase 2 (SHMT2), and Thymidylate Synthase (TYMS) (Figure 1).
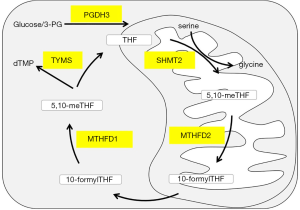
Single 1CM enzymes have been shown to play an important role in tumor initiation and progression. MTHFD2 catalyzes the transformation of methylene tetrahydrofolate and is strongly expressed in embryonic development, but is absent in most healthy adult tissues (10,11). Increased expression level of MTHFD2 is associated with poor prognosis of hepatocellular carcinoma, breast cancer and colorectal cancer (12-15). In addition to its role as a mitochondrial enzyme of the folic acid metabolism, nuclear localization close to newly synthesized DNA and a cell proliferation-enhancing effect of MTHFD2 were also found (16). Especially, the importance of MTHFD2 in lung cancer was manifested by modulation redox homeostasis and cell cycle progression (17,18). Furthermore, Takezawa et al. described that the increased expression of TYMS in highly proliferative tumor cells has been implicated in pemetrexed resistance in NSCLC (19).
Even though single factors have been evaluated in several cancer types, a comprehensive correlation between expression, proliferation and response towards chemotherapy in the three main lung cancer subgroups has not been conducted so far.
In this study we characterized the expression levels and clinical impact of the 1CM enzymes MTHFD2, PGDH3, SHMT2, MTHFD1 and TYMS in pulmonary AC, SQCLC and SCLC patient tissue samples. Furthermore, we functionally analyzed the 1CM enzymes according to cell proliferation and their role in the response towards pemetrexed and cisplatin in 15 lung cancer cell lines including all subgroups. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-1039).
Methods
Human tissue samples
Tissue samples were obtained from surgical resections at the Department of Thoracic and Cardiovascular Surgery of the University Medical Center, Göttingen. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the University Medical Center Göttingen (#1-2-08, 24-4-20) and informed consent was taken from all the patients.
Immunohistochemical staining
Tissue samples were assembled in tissue microarrays and IHC staining was performed as described before (20-22). Briefly, 2-µm tissue sections were incubated in EnVision Flex Target Retrieval Solution with either pH low or high (Dako). Tissue slices were incubated with primary antibody against MTHFD2 (dilution: 1:100, pH low, Abnova, #H00010797-M01), PGDH3 (1:500, pH low, Abcam, #AB57030), SHMT2 (1:200, pH low, Cell signaling, #37004), MTHFD1 (1:500, pH low, Atlas Antibodies, #HPA001290), TYMS (1:50, pH high, Abcam, #AB232021) at room temperature for 20 min. Sections were incubated with secondary antibody (EnVision Flex+, Dako) according to the manufacturer’s protocol and visualized through DAB (Dako). Slides were counterstained with Hematoxylin and staining were evaluated under light microscopy according to intensity: zero = negative staining; one = weak staining intensity; two = strong staining intensity.
Cell culture
H1993, H2228, HCC44, HCC78, HCC15, H2170, H520, EBC-1, HCC33, H1339, H82 human lung cancer cell lines were purchased from American Type Culture Collection (ATCC), DMS114, H3122 and H69 cell lines were obtained from the National Cancer Institute (NCI) of the National Institute of Health (NIH). EBC-1 KRAS cells with a heterozygous knock-in of a KRAS gene with an activating mutation was obtained from Horizon Discovery. Cells were maintained in RPMI-1640 (Gibco) supplemented with 10% fetal bovine serum, 1% Penicillin/Streptomycin and 1% L-Glutamine. Cells were incubated in humidified atmosphere of 5% CO2 at 37 °C. Medium was refreshed every 2–3 days and culture was passaged approximately at 80% confluency.
Transfection
All siRNAs were obtained from Qiagen, Hilden, Germany. Cells were transfected with 20 nM siRNA against either MTHFD2 (Nr. #1 SI02664928, #2 SI02664921), PGDH3 (Nr. #1 SI00090405, #2 SI05040994), SHMT2 (Nr. #1 SI04171237, #2 SI04176501), MTHFD1 (Nr. #1 SI02653084, #2 SI00083027) or TYMS (Nr. #1 SI02780757, #2 SI00021609). HiPerFect Transfection Reagent (Qiagen) was used according to the manufacturer’s protocol. Allstars negative siRNA was used as a scrambled control. Either a pcDNA3-MTHFD2 vector (CloneID: OHu18706, GenScript) or an empty pCDNA3 vector backbone (138209, Addgene) was transfected into AC cell lines for overexpression of MTHFD2. Cells expressing de-novo MTHFD2 were selected with G418 (at concentration of 700 µg/mL) for at least one week and MTHFD2 protein levels were confirmed by Western Blotting.
Western blotting
Cells were lysed with NP-40 buffer (1% NP40, pH 7.6, 150 mM NACL, 50 mM Tris, 1 mM Sodium orthovanadate, 1x Complete-EDTA, 0.2% Lauryl Maltoside) on ice for 30 min followed by centrifuging at 14,000 RPM at 4 °C for 20 min and protein content was determined using DCTM Protein Assay kit according to the manufacturer’s protocol. Proteins were immunoblotted as described before (23). Briefly, 20 µg protein were denatured at 95 °C for 5 min in Laemmli buffer, separated on a gradient polyacrylamide gel (4–15%) (BioRad) and transferred onto a nitrocellulose membrane with the Trans-Blot Turbo system (BioRad) according to manufacturer’s guideline. Membranes were blocked in 5% milk for 1 h following primary antibody exposure over night at 4 °C. The following antibodies and related secondary antibodies (DAKO) were use with a dilution of 1:1,000 in TBST for Anti-PGDH3 (#HPA021241, Sigma), Anti-SHMT2 (#HPA020549, Atlas Antibodies), Anti-MTHFD1 (#HPA001290, Atlas Antibodies), TYMS (#AB232021, Abcam) and Anti-MTHFD2 (#H00010797-M01, Abnova) were used. Anti-PARK7 (1:1,000, #AB76008, Abcam) was used as loading control (24).
Cell viability assay
For siRNA viability assay, 25 µL transfection medium containing 0.1 µL siRNAs/negative siRNA, 0.4 µL HiPerFect and 24.5 µL medium serum-free was incubated for 10 min at RT and added onto [1–5]×104 cells/well with 175 µL culture medium after seeding. For treatment viability assay, 2,500 cells/well were seeded in 96-well plate with 100 µL medium overnight and treated with the absence or presence of increasing concentrations of cisplatin (0.1, 1, 5, 10, 50, 100 µM) (Hexal AG, Holzkirchen, Germany) or pemetrexed (0.00632, 0.0125, 0.025, 1.5625, 3.125, 6.25, 12.5, 25, 50 µM) (Sigma-Aldrich, Taufkirchen, Germany) for indicated time as specified in the results. Cell proliferation was determined by MTS assay (Promega, Madison, WI, USA) according to the manufacturer’s protocol. Briefly, cells were incubated with 20 µL of CellTiter 96 Aqueous One Solution Reagent at 37 °C, 5% CO2 for 2 hours and 96-well plates were measured on a Tecan Reader Infinite 2000 using the i-control™ Microplate Reader Software at 490 and 650 nm.
Statistical analysis
Statistical analyses were performed using GraphPad Prim 8. Correlation between clinical parameters and 1CM enzymes expression was analyzed using chi-square test. Survival curves were drawn using Kaplan-Meier analyses and the significance was calculated by log-rank test. Students t-test was used for two group comparisons. More than two matched groups were analyzed using one-way ANOVA and Tukey’s multiple comparisons tests. Correlation between 1CM enzymes and half maximal inhibitory concentration (IC50) was performed by the Pearson’s correlation test. Statistical differences were considered significant at P<0.05.
Results
Correlation between expression of one-carbon metabolism enzymes and clinicopathologic characteristics
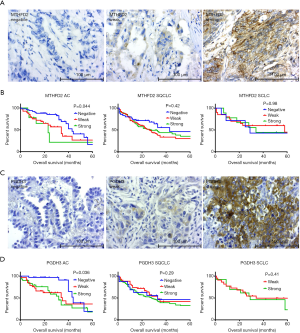
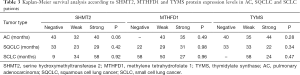
We performed immunohistochemical staining of the 1CM enzymes MTHFD2, PGDH3, SHMT2, MTHFD1 and TYMS in a patient cohort of in total 323 lung cancer patients (Figure 2 and Figure S1A). All patients were treated with surgical tumor resection without prior chemotherapy. The clinical characteristics of the patients are summarized in Table 1. Male patients (AC 57%, SQCLC 84%, SCLC 73%) were more frequent than female patients. The median age at time of diagnosis among the three groups was 66 years (range, 34–85 years). The majority of patients with AC (74%) and SQCLC (77%) showed a moderately differentiated disease. Frequency of T1-2 stage was 80%, 72% and 90% in AC, SQCLC and SCLC patients, respectively. Frequency of patients without lymph node metastasis (AC: 63%, SQCLC: 56%, SCLC: 75%) was higher than frequencies of N1, N2 and N3 together. Median follow-up time was 23, 26 and 34 months and death cases were reported in 48, 131 and 22 patients of AC, SQCLC and SCLC, respectively.
Full table
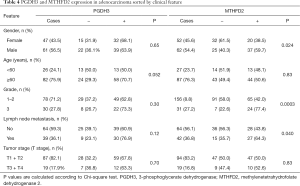
In SQCLC and SCLC, all 1CM enzymes meanwhile in AC only MTHFD1 and TYMS were expressed (either strong or weak) in more than 80% of cases. MTHFD2, SHMT2 and PGDH3 expression were significantly less abundant in AC with 50%, 63.4% and 65.7% respectively (Table 2). Only the expression of MTHFD2 and PGDH3 significantly correlated with a shorter overall survival only in AC (P=0.044 and P=0.036, respectively, Figure 2, Table 3). Furthermore, MTHFD2 expression was significantly associated with male gender, poorly differentiation grade and the occurrence of lymph node metastasis in AC patients (P=0.024, 0.0003 and 0.040, respectively, Table 4) in AC. Even though some factors showed a tendency to significance no other relevant difference or association to clinical parameters was detected (Figure S1B,C,D, and Tables 3,4).

Full table
Full table
Full table
Proliferation of pulmonary adenocarcinoma cells strongly depend on 1CM activity
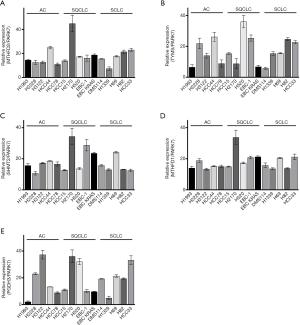
For a functional analysis of the 1CM proteins in vitro we used five AC cell line (H1993, H2228, H3122, HCC44 and HCC78), five SQCLC cell lines (HCC15, H2170, H520, EBC-1 and EBC-1 KRAS) and five SCLC cell lines (DMS114, H1339, H69, H82 and HCC33). Protein expression was evaluated by western blot and quantified over three replicates (Figure 3 and Figure S2). Meanwhile the expression of MTHFD1, MTHFD2 and SHMT2 was comparable in most of the cell lines, the expression of TYMS and PGDH3 demonstrated a higher variability. Interestingly, the expression of MTHFD2 and TYMS was significantly higher in the cell line HCC44 that harbors an activating KRAS mutation than in the other AC cell lines whereas SHMT2 was only moderately increased (Figure 3A,B,C).
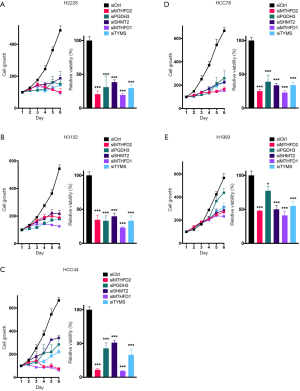
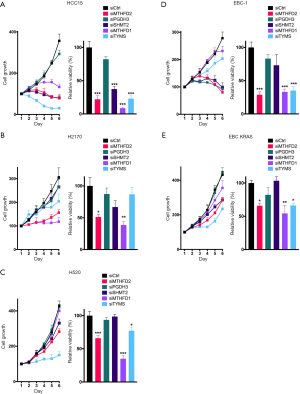
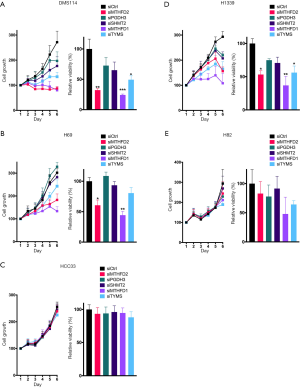
Next we aimed to investigate the effect of 1CM enzymes on cell proliferation using siRNAs that lead to a specific knockdown of the five described 1CM enzymes (Figure S3). The knockdown with two different siRNA against every single factor resulted in a significant decrease in cell proliferation in four of the five AC cell lines (Figure 4 and Figure S4). Especially in the KRAS mutated HCC44 with high MTHFD2 expression the inhibitory effect was stronger than 90%. In contrast, the reduction of the low expressed PGDH3 and TYMS in the AC cell line H1993 only moderately inhibited cell growth (less than 50%). In the SQCLC cell lines only the knockdown of MTHFD1 and MTHFD2 showed a constant reduction of cell proliferation and in SCLC there were only three cell lines (DMS114, H1339, H69) that showed a reduced cell growth after siRNA treatment against MTHFD1 and MTHFD2 (Figures 5,6).
MTHFD2 expression marks resistance to pemetrexed in AC cell lines
In order to evaluate the sensitivity of lung cancer cell lines to clinically used chemotherapeutics the described cell lines were treated with increasing concentrations of pemetrexed and cisplatin. Pemetrexed treatment resulted in a broad range of responses in the AC cell lines with IC50 levels between 0.027 to 113 µM (Figure 7A). Especially HCC44 showed a strong resistance to pemetrexed with an IC50 of 113 µM. Compared to the AC cell lines the SQCLC and the SCLC cell lines responded in a narrower range with an in average much higher IC50 (Figure 7B and Figure S5A). All cell lines showed a comparable response to cisplatin with IC50 ranging from 1.88 to 27.11 µM (Figure S5B,C,D). In comparison to the expression of one-carbon enzymes, MTHFD2 was the only enzyme that correlated to the response to pemetrexed in AC cell lines (r=0.87, P=0.03, Figure 7C). Although, there was a tendency for better response to pemetrexed treatment in MTHFD2 low expression SCLC cell lines (r=0.85, P=0.07, Figure 7D) there was not a significant correlation neither with pemetrexed nor with cisplatin (Figures S6,S7). However, we did not observe any further significant correlation between 1CM proteins expression and drug sensitivity, there was a tendency for better response to pemetrexed treatment in TYMS low expression AC and SCLC cells (r=0.27, P=0.06, and r=0.84, P=0.06, respectively, Figure S6A,B).
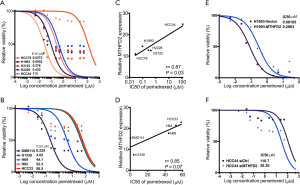
To explore the Pemetrexed resistance function of MTHFD2 in AC cells, we transfected a pcDNA-MTHFD2 or an empty vector or a specific siRNA against MTHFD2 or a non-target control siRNA in AC cells and treated the resulting cells with increasing concentrations of Pemetrexed. Overexpression of MTHFD2 in H1993 cells markedly increased viability of cells when treated with Pemetrexed compared to empty vector treated cells (IC50 for Pemetrexed: 0.30 and 0.081 µM, respectively, Figure 7E, Figure S8A,B). In contrast, MTHFD2 silencing significantly reduced cell viability after Pemetrexed treatment when compared to control treatment in HCC44 cells (IC50 Pemetrexed: 30.2 and 110.7 µM, respectively, Figure 7F). This indicated that resistance to Pemetrexed was induced by MTHFD2 overexpression and restored by its knockdown in AC cells.
Discussion
Increased 1CM and specifically MTHFD2, PGDH3 and TYMS activity has been associated with poor prognosis of cancer indicating the essential role of 1CM in saturating the need of newly synthesized nucleotides in proliferating cancer cells (25-29). Targeting 1CM metabolism with pemetrexed has been shown to be beneficial for patients with pulmonary adenocarcinoma (AC) (30). However, a comprehensive study of the main 1CM factors expression, function and correlation to chemoresistance in lung cancer was still missing.
In our cohort of lung cancer samples, expression of MTHFD2 and PGDH3 in AC group was significantly less common than in squamous cell lung cancer (SQCLC) or SCLC groups indicating a subtype specific diversity of 1CM activity in lung cancer. Previous studies have shown that MTHFD2 expression is correlated to worse prognosis of lung cancer, hepatocellular carcinoma, human pancreatic cancer, colorectal cancer and renal cell carcinoma patients (12,15,31-33). Similarly, PGDH3 has been proposed to be a prognostic factor in human breast cancer (14) and has been linked to tumorigenesis and poor prognosis in NSCLC (25). Through inclusion of the three major lung cancer subgroups, we could pinpoint that expression of MTHFD2 and PGDH3 was correlated to worse prognosis particularly in AC but not in SQCLC or SCLC patients. In addition, MTHFD2 expression correlated significantly with male gender, poor differentiation grade and lymph node metastasis in AC patients, suggesting an association between MTHFD2 expression and AC disease progression.
1CM enzymes have been described to fulfill the cancer-specific nutrient demand in tumor cell proliferation (28,29,34). Proliferation of AC cell lines was more affected upon siRNA knockdown of MTHFD2, PGDH3, SHMT2, MTHFD1 and TYMS than SQCLC or SCLC cell lines. In accordance to these results the down-regulation of MTHFD2 in NSCLC was shown to inhibit cell proliferation via regulating cell cycle arrest and redox homeostasis (17,18). Dependency of cancer cells proliferation on 1CM was also observed in breast cancer, colorectal cancer (16,35), Ewing sarcoma (36), human hepatocellular carcinoma (13,37) and glioma (38).
MTHFD2 and PGDH3 protein levels in the SQCLC cell line H2170 and the SCLC cell line HCC33 were significantly higher than all other cell lines, however, there was no significant impact on cells viability upon siRNA knock down. In summary, according to protein expression levels and functional analyses of 1CM enzymes, we propose a stronger dependency on 1CM activity in AC compared to SQCLC or SCLC.
We observed a stronger expression of MTHFD2, SHMT2 and TYMS in the KRAS mutant cell line HCC44 in comparison to KRAS wild type cell lines. This goes along with recent studies, which have described a higher dependency on folate metabolism in KRAS mutated NSCLC cells (39). Here, we show that four AC cell lines with low levels of MTHFD2 expression exhibited higher response to pemetrexed in comparison to HCC44 with high levels of MTHFD2 expression. We furtherly confirmed the correlation between MTHFD2 expression and resistance to Pemetrexed by showing that overexpression of MTHFD2 leads to increased resistance to Pemetrexed meanwhile knockdown of MTHFD2 reduces resistance to Pemetrexed. Accordingly, Nishimura et al. has described that MTHFD2 increased chemotherapy resistance to Gefitinib in pulmonary adenocarcinoma (40). The SQCLC and SCLC cell lines H2170, H520, H82 and HCC33 showed not only the highest expression of MTHFD2 but also the strongest resistance against pemetrexed. These results suggest that high expression of MTHFD2 could shape a primary indicator of treatment resistance against pemetrexed. Moreover, our findings provide additional evidences that high expression of MTHFD2 not only correlates with worse prognosis of AC patients, but also contributes to the sensitivity to Pemetrexed. Therefor MTHFD2 might be a useful biomarker for the decision of a specific treatment strategy in AC.
A persuasive MTHFD2 inhibitor (DS18561882) has already been characterized to have an anti-tumor efficacy in vivo in human breast cancer (41). Taking into consideration that MTHFD2 is only expressed in developmental and neoplastic cells and is almost absent in healthy adult tissues (11) makes MTHFD2 a potential new therapeutic option in AC with further need for validation.
Although the correlative analyses with TYMS expression and Pemetrexed resistance did not produce a statistically significant correlation, there was also a trend for better response to pemetrexed treatment in TYMS low expression AC and SCLC cell lines (r=0.27, P=0.06 and r=0.84, P=0.07, respectively, Figure S6A,B). These findings support previous studies, which suggested that TYMS was a predictive factor for sensitivity to pemetrexed in lung cancer (19,42,43).
Overall, our study revealed a diversity of 1CM enzyme expression and effects in human lung cancer subtypes. In pulmonary adenocarcinoma, MTHFD2 might be both a predictive biomarker that could help for chemotherapy selection and a potential new drug target especially as MTHFD2 is almost not expressed in healthy adult tissue.
Acknowledgments
DMS114, H3122 and H69 human lung cancer cell lines are from the National cancer institute (NCI) of the National Institute of Health (NIH). We thank Jennifer Appelhans for her technical support. We acknowledge support by the Open Access Publication Funds of the Göttingen University.
Funding: HB and OE are supported by the Deutsche Krebshilfe Foundation (grant: 70112551). HB is supported by the University medical center Göttingen and the Else-Kröner-Fresenius-Foundation. SY and LP are supported by the Chinese Scholarship Counsil.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-1039
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-1039
Peer Review File: Available at http://dx.doi.org/10.21037/tlcr-20-1039
Conflict of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-1039). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the University Medical Center Göttingen (#1-2-08, 24-4-20) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- World Health Organization: GLOBOCAN 2008 - Estimated cancer Incidence, Mortality, Prevalence and Disability-adjusted life years (DALYs) Worldwide in 2008. Available online: http://globocan.iarc.fr/
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. Erratum in: Nature 2014;514:262 Rogers, K [corrected to Rodgers, K]. Erratum in: Nature 2018;559:E12. [Crossref] [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Papadopoulos KP, Ou SHI, Johnson ML, et al. A phase I/II multiple expansion cohort trial of MRTX849 in patients with advanced solid tumors with KRAS G12C mutation. J Clin Oncol 2019;37:TPS3161 [Crossref]
- Scagliotti GV, Parikh P, von Pawel J, et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol 2008;26:3543-51. [Crossref] [PubMed]
- Ducker GS, Rabinowitz JD. One-Carbon Metabolism in Health and Disease. Cell Metab 2017;25:27-42. [Crossref] [PubMed]
- Tibbetts AS, Appling DR. Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu Rev Nutr 2010;30:57-81. [Crossref] [PubMed]
- Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer 2013;13:572-83. [Crossref] [PubMed]
- Mejia NR, MacKenzie RE. NAD-dependent methylenetetrahydrofolate dehydrogenase is expressed by immortal cells. J Biol Chem 1985;260:14616-20. [Crossref] [PubMed]
- Nilsson R, Jain M, Madhusudhan N, et al. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun 2014;5:3128. [Crossref] [PubMed]
- Liu X, Huang Y, Jiang C, et al. Methylenetetrahydrofolate dehydrogenase 2 overexpression is associated with tumor aggressiveness and poor prognosis in hepatocellular carcinoma. Dig Liver Dis 2016;48:953-60. [Crossref] [PubMed]
- Lee D, Xu IM, Chiu DK, et al. Folate cycle enzyme MTHFD1L confers metabolic advantages in hepatocellular carcinoma. J Clin Invest 2017;127:1856-72. [Crossref] [PubMed]
- Liu F, Liu Y, He C, et al. Increased MTHFD2 expression is associated with poor prognosis in breast cancer. Tumour Biol 2014;35:8685-90. [Crossref] [PubMed]
- Ju HQ, Lu YX, Chen DL, et al. Modulation of Redox Homeostasis by Inhibition of MTHFD2 in Colorectal Cancer: Mechanisms and Therapeutic Implications. J Natl Cancer Inst 2019;111:584-96. [Crossref] [PubMed]
- Gustafsson Sheppard N, Jarl L, Mahadessian D, et al. The folate-coupled enzyme MTHFD2 is a nuclear protein and promotes cell proliferation. Sci Rep 2015;5:15029. [Crossref] [PubMed]
- Yu C, Yang L, Cai M, et al. Down-regulation of MTHFD2 inhibits NSCLC progression by suppressing cycle-related genes. J Cell Mol Med 2020;24:1568-77. [Crossref] [PubMed]
- Chan CH, Wu CY, Dubey NK, et al. Modulating redox homeostasis and cellular reprogramming through inhibited methylenetetrahydrofolate dehydrogenase 2 enzymatic activities in lung cancer. Aging (Albany NY) 2020;12:17930-47. [Crossref] [PubMed]
- Takezawa K, Okamoto I, Okamoto W, et al. Thymidylate synthase as a determinant of pemetrexed sensitivity in non-small cell lung cancer. Br J Cancer 2011;104:1594-601. [Crossref] [PubMed]
- Bohnenberger H, Kaderali L, Strobel P, et al. Comparative proteomics reveals a diagnostic signature for pulmonary head-and-neck cancer metastasis. EMBO Mol Med 2018;10:e8428 [Crossref] [PubMed]
- Buentzel J, Yao S, Elakad O, et al. Expression and prognostic impact of alpha thalassemia/mental retardation X-linked and death domain-associated protein in human lung cancer. Medicine (Baltimore) 2019;98:e16712 [Crossref] [PubMed]
- Bremer SCB, Conradi LC, Mechie NC, et al. Enhancer of Zeste Homolog 2 in Colorectal Cancer Development and Progression. Digestion 2021;102:227-35. [PubMed]
- Elakad O, Lois AM, Schmitz K, et al. Fibroblast growth factor receptor 1 gene amplification and protein expression in human lung cancer. Cancer Med 2020;9:3574-83. [Crossref] [PubMed]
- Wiśniewski JR, Mann M. A Proteomics Approach to the Protein Normalization Problem: Selection of Unvarying Proteins for MS-Based Proteomics and Western Blotting. J Proteome Res 2016;15:2321-6. [Crossref] [PubMed]
- Zhang B, Zheng A, Hydbring P, et al. PHGDH Defines a Metabolic Subtype in Lung Adenocarcinomas with Poor Prognosis. Cell Rep 2017;19:2289-303. [Crossref] [PubMed]
- DeNicola GM, Chen PH, Mullarky E, et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet 2015;47:1475-81. [Crossref] [PubMed]
- Jain M, Nilsson R, Sharma S, et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 2012;336:1040-4. [Crossref] [PubMed]
- Locasale JW, Grassian AR, Melman T, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet 2011;43:869-74. [Crossref] [PubMed]
- Possemato R, Marks KM, Shaul YD, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 2011;476:346-50. [Crossref] [PubMed]
- Tomasini P, Barlesi F, Mascaux C, et al. Pemetrexed for advanced stage nonsquamous non-small cell lung cancer: latest evidence about its extended use and outcomes. Ther Adv Med Oncol 2016;8:198-208. [Crossref] [PubMed]
- Director's Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med 2008;14:822-7. [Crossref] [PubMed]
- Song Z, Feng C, Lu Y, et al. PHGDH is an independent prognosis marker and contributes cell proliferation, migration and invasion in human pancreatic cancer. Gene 2018;642:43-50. [Crossref] [PubMed]
- Lin H, Huang B, Wang H, et al. MTHFD2 Overexpression Predicts Poor Prognosis in Renal Cell Carcinoma and is Associated with Cell Proliferation and Vimentin-Modulated Migration and Invasion. Cell Physiol Biochem 2018;51:991-1000. [Crossref] [PubMed]
- Mullarky E, Mattaini KR, Vander Heiden MG, et al. PHGDH amplification and altered glucose metabolism in human melanoma. Pigment Cell Melanoma Res 2011;24:1112-5. [Crossref] [PubMed]
- Chen J, Chung F, Yang G, et al. Phosphoglycerate dehydrogenase is dispensable for breast tumor maintenance and growth. Oncotarget 2013;4:2502-11. [Crossref] [PubMed]
- Sen N, Cross AM, Lorenzi PL, et al. EWS-FLI1 reprograms the metabolism of Ewing sarcoma cells via positive regulation of glutamine import and serine-glycine biosynthesis. Mol Carcinog 2018;57:1342-57. [Crossref] [PubMed]
- Woo CC, Chen WC, Teo XQ, et al. Downregulating serine hydroxymethyltransferase 2 (SHMT2) suppresses tumorigenesis in human hepatocellular carcinoma. Oncotarget 2016;7:53005-17. [Crossref] [PubMed]
- Zhao M, Tan B, Dai X, et al. DHFR/TYMS are positive regulators of glioma cell growth and modulate chemo-sensitivity to temozolomide. Eur J Pharmacol 2019;863:172665 [Crossref] [PubMed]
- Moran DM, Trusk PB, Pry K, et al. KRAS mutation status is associated with enhanced dependency on folate metabolism pathways in non-small cell lung cancer cells. Mol Cancer Ther 2014;13:1611-24. [Crossref] [PubMed]
- Nishimura T, Nakata A, Chen X, et al. Cancer stem-like properties and gefitinib resistance are dependent on purine synthetic metabolism mediated by the mitochondrial enzyme MTHFD2. Oncogene 2019;38:2464-81. [Crossref] [PubMed]
- Kawai J, Toki T, Ota M, et al. Discovery of a Potent, Selective, and Orally Available MTHFD2 Inhibitor (DS18561882) with in Vivo Antitumor Activity. J Med Chem 2019;62:10204-20. [Crossref] [PubMed]
- Christoph DC, Asuncion BR, Hassan B, et al. Significance of folate receptor alpha and thymidylate synthase protein expression in patients with non-small-cell lung cancer treated with pemetrexed. J Thorac Oncol 2013;8:19-30. [Crossref] [PubMed]
- Agulló-Ortuño MT, Garcia-Ruiz I, Diaz-Garcia CV, et al. Blood mRNA expression of REV3L and TYMS as potential predictive biomarkers from platinum-based chemotherapy plus pemetrexed in non-small cell lung cancer patients. Cancer Chemother Pharmacol 2020;85:525-35. [Crossref] [PubMed]


