
Mitogen signal-associated pathways, energy metabolism regulation, and mediation of tumor immunogenicity play essential roles in the cellular response of malignant pleural mesotheliomas to platinum-based treatment: a retrospective study
Introduction
Malignant pleural mesothelioma (MPM) is a malignancy originating from pleural mesothelium, a serosal membrane covering the thoracic cavity. This rare type of cancer is associated with asbestos exposure and has a very poor prognosis: less than 20 months if treated and less than nine months if untreated (1-3). Only a small fraction of patients is suitable for potentially curative surgery through radical pleurectomy (4). Despite encouraging results of the recent trials with immunotherapy in patients with MPM (5), most patients receive the standard chemotherapy, approved by both the U.S. Food and Drug Administration (FDA) as well as the European Medicines Agency (EMA). This treatment, consisting of cisplatin and pemetrexed, prolongs overall survival by several months, with especially epithelioid mesothelioma benefiting from this therapy (6). Response rates for monotherapies either with cisplatin or carboplatin are low, ranging from 6% to 16% (7,8).
Platinum cytotoxicity is based on the formation of bulky DNA adducts induced by covalent binding of platinum and alteration of DNA bases (9), leading to both DNA inter- and (1,2 or 1,3) intra-strand cross-linking (10-17). Platinum compounds prevent normal cell replication and trigger apoptosis (12,16,18) unless adducts from genomic DNA are repaired (15). Even a single DNA cross-link, if not repaired, can be lethal (19). Platinum-induced DNA damage leads to TP53 induced cell cycle arrest and apoptosis (20,21). However, it is uncertain whether DNA repair mechanisms are the key factors associated with an impaired therapy response since other cellular mechanisms, such as cell cycle control and intracellular signaling pathways, are thought to contribute to resistance to platinum. Since the identification of MPM molecular properties may help to identify the reason(s) of the inadequate treatment response, several studies addressed this question (7,8,19,22-25). Unfortunately, the reasons for the poor efficacy of platinum-based therapy in MPM patients are still largely unknown. Given these therapeutic limitations, further basic research is needed to provide a more detailed insight into the pathogenesis and biology of MPM and enable opportunities for innovative and novel treatment strategies (1).
Therefore, we aimed to analyze differences in key signaling pathways and biological mechanisms in therapy-naïve samples compared to samples after chemotherapy to evaluate the effect of platinum-based chemotherapy.
We present the following article in accordance with the STROBE reporting checklist (available at https://dx.doi.org/10.21037/tlcr-21-201).
Methods
Study cohort and experimental design
For this exploratory mRNA study, twenty-four formalin-fixed paraffin-embedded (FFPE) tumor specimens of MPM patients of the epithelioid histological subtype treated at the West German Cancer Centre or the West German Lung Centre between 2005 and 2009 were screened. Diagnoses of MPM, epithelioid subtype, were confirmed by two experienced pathologists (TMA, JWO), based on the latest WHO classification (26), and patients were re-staged using the UICC/AJCC staging system of 2017 (27). Half of the collected samples were taken prior to any therapy, and the other half after chemotherapy. All patients received platinum-based chemotherapy. The radiologic response rate was assessed using the modified Response Evaluation Criteria in Solid Tumors (modRECIST), which have been validated in MPM (28,29). Surveillance for this study cohort stopped on August 31, 2014. Complete follow-up was available for all patients with reported deaths in more than 96% (23/24). Disease progression under therapy was observed in approximately 80% (19/24) of patients. Clinico-pathological data are summarized in Table 1.

Full table
This retrospective study conformed to the principles outlined in the Declaration of Helsinki (as revised in 2013). It was approved by the Ethics Committee of the Medical Faculty of the University Duisburg-Essen (identifier: 14-5775-BO), and individual consent for this retrospective analysis was waived.
RNA extraction and RNA integrity assessment
Three to five 4 µm thick paraffin sections of whole tumor per sample were used for total RNA isolation. RNA was isolated by a semi-automatic Maxwell purification system (Maxwell RSC RNA FFPE Kit, AS1440, Promega). Purification was performed according to the manufacturer’s instructions. RNA was eluted in 50 µL RNase-free water and stored at −80 °C. RNA concentration was measured using a Qubit 2.0 fluorimeter (Life Technologies) appertaining to the RNA broad-range assay. Total RNA integrity and quality were assessed using a Fragment Analyzer (Advanced Analytical Technologies, Ames, IA, USA) and appertaining DNF-489 standard sensitivity RNA analysis kit.
Digital gene expression analysis
Gene expression patterns were screened for prognostic and predictive biomarkers using the NanoString nCounter platform for digital gene expression analysis with an appurtenant custom-designed panel comprising a total of 366 mRNAs (9 potential reference genes (ACTB, GAPDH, HPRT1, CLTC, GUSB, NEDD8, PGK1, TTC1 and TUBB as well as 357 target genes) covering the most important tumor signaling pathways. For each sample, 200 ng of total RNA was processed. The sample preparation in the nCounter Prep Station (NanoString) was carried out by using the high-sensitivity protocol (3 h preparation). The cartridges were measured at 555 fields of view in the nCounter Digital Analyzer (NanoString).
NanoString data processing
NanoString data processing was performed with the R statistical programming environment (v3.4.2) using the NanoStringNorm (30) and the NAPPA package, respectively.
Considering the counts obtained for positive control probe sets raw NanoString counts for each gene were subjected to a technical factorial normalization, carried out by subtracting the mean counts plus two-times standard deviation from the CodeSet inherent negative controls. Afterward, a biological normalization using the geometric mean of the inherent reference genes was performed. Additionally, all counts with P>0.05 after one-sided t-test versus negative controls plus 2× standard deviations were interpreted as not expressed to overcome basal noise.
Statistical analysis
Statistical and graphical analyses were performed with the R statistical programming environment (v4.0.2) (31). Before explorative data analysis, the Shapiro-Wilks-test was applied to test for normal distribution of each data set. For dichotomous variables, the Wilcoxon Mann-Whitney rank sum test (non-parametric) or two-sided student’s t-test (parametric) was used. For ordinal variables with more than two groups, the Kruskal-Wallis test (non-parametric) or ANOVA (parametric) was used to detect group differences. Double dichotomous contingency tables were analyzed using Fisher’s exact test. To test dependence to ranked parameters with more than two groups, Pearson’s Chi-squared test was used. Correlations between metrics were analyzed using Spearman’s rank correlation test.
Adaption of profiles for diagnostic purposes was explored with the supervised machine learning tools “Classification and Regression Tree Algorithm” (CART) as implemented in the “rpart” library of R (according to Therneau and Atkinson) using a leave-one-out cross-validation. Additionally, conditional interference trees (CTree), as implemented in the “party” library of R was modeled, leading to a non-parametric class of tree-structured regression models.
Pathway analysis is based on the KEGG database and was performed using the “pathview” package of R. Differences were specified by –log2 fold change between means (if parametric) or medians (if non-parametric) of compared groups. Significant pathway associations were identified by gene set enrichment analysis using the WEB-based GEne SeT AnaLysis Toolkit (WebGestalt) (31-33). Each run was executed with 1,000 permutations. Finally, all associations were ranked according to the false discovery rate (FDR) (P<0.05).
Due to the multiple statistical tests, the P values were adjusted by using the FDR. The level of statistical significance was defined as P≤0.05 after adjustment.
Results
Gene expression analysis
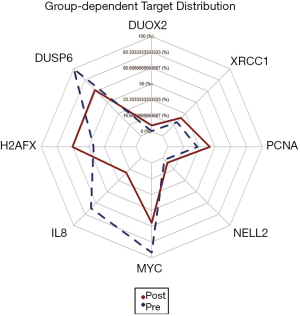
Tumor specimens collected after therapy show clearly elevated expression levels of H2AFX (P=0.005), PCNA (P=0.024), XRCC1 (P=0.029), DUOX2 (P=0.034) and NELL2 (P=0.034) in comparison to treatment-naïve samples. In contrast, gene expression of DUSP6 (P=0.010), MYC (P=0.020), IL8 (P=0.045) and ANGPT2 (P=0.050) was found to be significantly decreased after treatment (Figure 1).
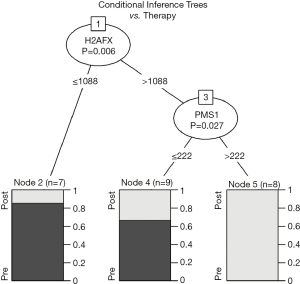
Additionally, unsupervised conditional inference tree (CTree) analysis identifies H2AFX and PMS1 as a pattern for biunique classification between neo- and adjuvant treatment. In particular, H2AFX expression below 1,088 counts leads to a subgroup highly enriched for pretreatment samples (P=0.006). Subsequently, a group with more than 1,088 H2AFX counts and high expression of PMS1 (P=0.027, cut-off: 222 counts) contains only specimens after platinum-based therapy (Figure 2).
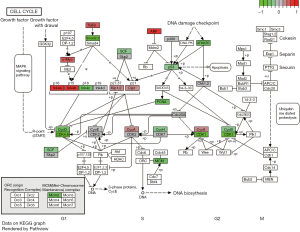
Looking into the factors involved in cell cycle control and progression in the pretreatment samples revealed high levels of TGF-β, p14, p16 (both encoded by CDKN2A), and p15 (encoded by CDKN2B), indicating a robust regulatory mechanism for G1-to-S-phase transition. In samples collected after platinum-based therapy, an upregulation of the following genes was observed: SMAD2 and SMAD3, markers for apoptosis/cell cycle arrest induction, ATM/ATR and their downstream cascade including p53 and PCNA, and also various CDKs such as CDK1, CDK4, and CDK6 as well as their respective cyclins B, D, and H (Figure 3).
Gene set enrichment analysis (GSEA)
GSEA was performed (Table S1) to identify affected biological processes. GSEA utilizes molecular interaction networks outlined by the Kyoto Encyclopedia of Gene and Genomes (KEGG) to map out increased gene expression in a specific molecular pathway, depending on a response variable (treatment) (Figures S1,S2).
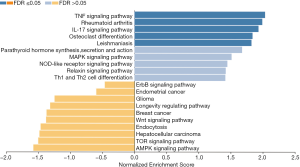
AMPK signaling pathway (normalized enrichment score: −1.58), mTOR signaling pathway (normalized enrichment score: −1.50), hepatocellular carcinoma (normalized enrichment score: −1.48), endocytosis (normalized enrichment score: −1.46), Wnt signaling pathway (normalized enrichment score: −1.38), breast cancer (normalized enrichment score: −1.37), longevity regulating pathway (normalized enrichment score: −1.31), glioma (normalized enrichment score: −1.25), endometrial cancer (normalized enrichment score: −0.59) and ErbB signaling pathway (normalized enrichment score: −0.46) are enriched in samples after platinum therapy. On the other hand, activity of TNF signaling pathway (normalized enrichment score: 2.03), rheumatoid arthritis (normalized enrichment score: 1.99), IL-17 signaling pathway (normalized enrichment score: 1.93), osteoclast differentiation (normalized enrichment score: 1.84), Leishmaniasis (normalized enrichment score: 1.82), parathyroid hormone synthesis, secretion and action (normalized enrichment score: 1.68), MAPK signaling pathway (normalized enrichment score: 1.51), NOD-like receptor signaling pathway (normalized enrichment score: 1.45), relaxin signaling pathway (normalized enrichment score: 1.42) and Th1 and Th2 cell differentiation (normalized enrichment score: 1.42) are associated with specimens collected from therapy-naïve patients (Figure 4).
Gene expression and clinical stage
Eight genes were identified to be differentially expressed depending on MPMs’ UICC stage. MAPK6 (P=0.007, rho=0.75), ICAM5 (P=0.041, rho=0.62) and ICAM4 (P=0.050, rho=0.60) show low expression levels in UICC 1A and B cases, increasing and reaching its plateau in higher UICC stages 3A, 3B and 4, where no differences in expression can be shown. Similarities can be observed for PARP1 (P=0.007, rho=0.74), DUOX2 (P=0.023, rho=0.67), SLCO3A1 (P=0.023, rho=0.67) and BRCA1 (P=0.041, rho=0.63) expression levels, showing a continuous rise from low expression in UICC 1A/B, rising from UICC 3A to 3B with higher gene expression to outstanding expression in UICC stage 4. For HGF (P=0.050, rho=0.60), the increased expression could only be detected in UICC stage 4.
Gene expression and stratification for overall survival (OS) and progression-free survival (PFS)
Gene expression levels of 22 targets have been identified to impact OS. Longer OS is associated with lower expression of 19 genes: ICAM5 (P=0.002), CD82 (P=0.004), FYN (P=0.010), WNT1 (P=0.011), FGF1 (P=0.013), CDK5R2 (P=0.015), MMP8 (P=0.019), IGSF5 (P=0.033), SLC7A6 (P=0.033), SLIT1 (P=0.036), SLCO1B3 (P=0.037), WISP3 (P=0.039), MLH1 (P=0.040), GLUT1 (P=0.043), IGSF5 (P=0.043), TCEB2 (P=0.048) and claudins, including CLDN14 (P=0.004), CLDN3 (P=0.010) and CLDN11 (P=0.011). In addition, CLDN2 (P=0.055) and CLDN5 (P=0.057) closely failed significance but show a clear trend for prolonged survival at low gene expression levels. On the other hand, expression of CDKN2A (P=0.022) and high expression of TIAM1 (P=0.024) and JAM2 (P=0.039) are detected in patients with longer OS (Figure 5).
Applying unsupervised, decision-tree-based machine learning (COX-CIT), the expression of MMP9 can stratify patients into three different risk groups, with different OS. Using two cut-offs (609 counts, P=0.004, and 1,492 counts, P=0.001) it can identify the patients with the worst prognosis (Node 4) (Figure 6A).
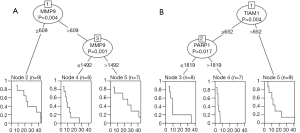
Applying the same COX-CIT algorithm for PFS, we were able to identify patients showing progression within the first 10 months (node 4), presenting low TIAM1 (P=0.004, cut-off 652 counts) and high PARP1 (P=0.017, cut-off: 1,819 counts) expression. Furthermore, the low TIAM1 and PARP1 expression group comprised another third of patients showing progression within the first year in up to 90% of cases. The remaining samples with high TIAM expression show better PFS times and rates, with a median survival of about one year, while one-fifth of patients show progression foremost after two years (Figure 6B).
Discussion
Chemoresistance, either intrinsic or acquired, substantially impairs the efficacy of chemotherapy, escalating mortality rates in cancer patients. Acquirement of resistance towards chemotherapeutics is a dynamic and multifactorial process influenced by cancer stem cell (CSC) enrichment (34). Several mechanisms leading to platinum resistance have been identified: reduced intracellular cisplatin accumulation due to alterations in transmembrane transport, activation of cell growth-promoting and DNA damage repair pathways, aberrant DNA methylation, enhanced epithelial-to-mesenchymal transition, and reduced endocytosis of cisplatin (35-37). However, as there are no established tools for predicting response to platinum therapy, there is an unmet need for predictive factors helping to identify patients at high risk for platinum resistance to select the adequate treatment for individual patients (37). Understanding the underlying mechanisms of cellular response to platinum may help to overcome this problem.
In the present study, we identified downregulation of MAPK-, IL-17-, relaxin- and TNF signaling pathways, and upregulation of AMPK-, mTOR-, Wnt- signaling and longevity regulation pathways in tumor samples of patients with MPM after platinum-based therapy.
Mitogen-activated protein kinase (MAPK)
MAPK signaling is important for cancer resistance to chemotherapy (38). Especially the p38-MAPK pathway seems to be involved in cisplatin-based chemotherapy response. In head and neck carcinoma, its low activity was in correlation to platinum resistance (39). In contrast, constitutive activation of the C-KIT/MAPK/MEK axis has been shown to result in a resistant phenotype of ovarian cancer cells treated with platin compounds (40).Furthermore, a broad miRNA screening study in vitro identified the miRNAs miR-20b*, let-7a, miR-524-3p, miRPlus-F1147, miR-300, miR-1299, miR-193b, miRPlus-F1064 as well as miR-642 as differentially expressed between sensitive and resistant ovarian cancer cell lines (41). Those miRNAs manipulate, among others, MAPK-, Wnt-, mTOR- as well as TGF-β-signaling (41). Of interest, also an RAF independent activation of MAPK cascade was proven to contribute to platin resistance in human cancer cells in vitro and in vivo (42). Taking all this into account, activation of the MAPK pathway is a prominent mechanism for platinum resistance in MPM; therefore, its downregulation in our samples after platinum-based chemotherapy corresponds most likely to the direct effect of the therapy.
Interleukin-17 (IL-17)
Since inflammation is present in most cancers, it is evident that different cytokines, like TNF-α, TGF-β, macrophage migratory inhibitory factor, and interleukins, are involved in promoting or inhibiting malignant growth, both in vitro and in vivo (43). Among those, the IL-17 family has a significant role in cancer immunity. Th17 (CD4+ T cells), innate tissue-resident cells (ITRC), and CD8+ cytotoxic T lymphocytes (CTLs) have been reported to participate in IL-17 production (44,45). IL-17 signaling in colorectal carcinoma promotes tumorigenesis by preventing the production of T cell-attracting CXCL9 and CXCL10 in tumor cells (46). Interestingly, asbestos exposure increases IL-17 production, especially in CD4+ surface CXCR3+ cells, indicating that the relation between Treg and Th17 fractions is important for asbestos immunogenicity and decrease in anti-tumor immunity (47). This is in accordance with the findings of Zebedeo et al., who found increased production of IL-17, IL-6, TGF-β, and TNF-α in mice following exposure to erionite and amphibole asbestos (48). Furthermore, IL17A mRNA expression and IL17A+ intratumoral cells in gastric cancer were predictive for better chemotherapy response (49). Moreover, in platinum-treated ovarian cancer, GSEA revealed enrichment in MAPK, ERBB, TNF, and IL‐17 signaling pathways in chemotherapy-sensitive patients (50). Besides the known pro-tumorigenic functions of IL-17, its increased presence is also a potential marker for chemotherapy sensitivity. Our finding of IL-17 pathway downregulation in MPM patients after platinum-based therapy is best explained as a platinum anti-tumorigenic effect.
Relaxin
Another pathway found to be downregulated in MPM samples after therapy is the relaxin-pathway. Relaxin is an anti-fibrotic agent. It hinders cytokine and growth-factor induced proliferation of fibroblasts and matrix production but also induces matrix degradation through increased activity of matrix‐degrading matrix metalloproteinases (MMP) (51,52). Its involvement in ECM remodeling, but also in angiogenesis, blood flow, anti-apoptosis, cell migration, and anti‐inflammation, helps in the proliferation, invasiveness, and metastasis of tumor cells and is highly expressed on them (51,53,54). Our finding of relaxin-pathway downregulation demonstrates the effect of platinum-based chemotherapy on this pro-tumorigenic factor.
TNF and NF-κB
Asbestos induces expression of TNF-α receptor I and secretion of TNF-α secretion in mesothelial cells (55). Furthermore, in mesothelial cells, TNF-α prevents apoptosis and cell death through NF-kappaB activation, reducing asbestos toxicity, allowing them to survive and go through malignant transformation (55,56). Activated NF-κB leading to upregulated PIK3CA expression via TNF-α has been identified as one of the key molecular features behind the action of cisplatin (34). Recent data indicates the PI3K/AKT pathway as decisive of cisplatin action in resistant cells (34). Platinum compounds have been shown to inhibit TNF-α stimulated NF-κB activation (57). This explains the downregulation of the TNF-pathway associated with platin-based treatment as a rather direct effect of therapeutic intervention.
AMPK and mTOR
AMP-activated protein kinase (AMPK) is a stress-response molecule involved in maintaining energy homeostasis in eukaryotic cells (58,59). Together with the mTOR downstream pathway, it regulates cellular metabolism, energy homeostasis, and cell growth (58). AMPK with LKB1 and P53 modulate mTOR and Akt signaling, resulting in cell growth inhibition and cell cycle arrest (60). It is known that the mTOR pathway is hyperactivated in many cancers (61-63). The PI3K/AKT/mTOR pathway has an important role in MPM tumorigenesis, and its blockade is therapeutically relevant, although it might induce rebound AKT activation leading to resistance (64). Interestingly, platinum-based chemotherapy did not prove effective against tumor cells with a hyperactivated mTOR pathway (65,66). Therefore dual inhibition of PI3K (for example, with cisplatinum), and mTOR might be a better option (67). Several studies indicate that AMPK activators inhibit the functions of myeloid-derived suppressor cells (MDSCs) and induce anti-tumor activities in many cancers (68-71). This is based on the fact that AMPK signaling inhibits immune signaling pathways, like JAK-STAT, NF-κB, C/EBPβ, CHOP, and HIF-1α, which are activating immunosuppressive MDSCs, therefore enabling adequate immune surveillance of tumor cells (68). In contrast, AMPK also has a pivotal role in chemoresistance (60).Evidence suggests that beyond classical resistance mechanisms, microenvironment stress also results in antagonism-independent alterations of the drug target, overactive DNA repair and survival pathways, enhanced expression of detoxification proteins, and drug efflux (72-74). It was shown that esophageal squamous carcinoma cells under nutrient stress, with activated AMPK pathway, kept proliferating and demonstrated chemoresistance to cisplatin (72). Both AMPK and mTOR pathways were upregulated in our samples after chemotherapy, most probably leading to the development of resistance to therapy.
Wnt
The interaction between the immune system and cancer is not fully understood but appears to be relevant for therapeutic decisions and response to platinum-based chemotherapy. While immune cells like CTLs and natural killer cells are well known for influencing outcome after chemotherapy in various cancers, also “non-immune” cells such as cancer cells or associated fibroblasts have an important role (75,76). They release molecules supporting angiogenesis, interact with the immune system, support tumor progression, induce degradation of the extracellular matrix, thereby enabling tissue invasion and metastasis (77-80). One potential inducer of TGF-β and VEGF are Wnt ligands, which can be produced, among others, by pulmonary fibroblasts. In non-small cell lung carcinoma, increased Wnt activity is associated with a higher risk for relapse (81-83). It has been demonstrated that Wnt ligands are potent modulators of the immune response and can suppress T cell activity (84-87) with some tumors producing them as an evasion strategy (88). Inhibition of Wnt signaling with cationic cyclometalated platinum(ii) complexes demonstrated efficient prevention of cancer cell proliferation and migration (89). Like the AMPK- and mTOR-, Wnt-pathway is also upregulated in our treated patient cohort, resulting in the development of chemotherapy-resistant tumor cells.
The interpretation of the results obtained in this study concerning different signaling pathways is challenging. Due to their multiple functions, it does not appear to be straightforward at first sight. Additionally, some effects can be assigned to cellular protection against the incoming oxidative cell stress, whereas some are related to mechanisms involved in overcoming cell longevity and progression. However, we have found downregulation in pathways important for carcinogenesis, which might be interpreted as the direct effect of the platinum-based therapy. Simultaneously, there is an upregulation in pathways involved in the resistance to this therapy, which eventually leads to disease progression.
There are several limitations to this study: it is a retrospective study, with a rather small cohort. Furthermore, we did not analyze paired samples from patients before and after therapy. However, our results are to be interpreted as a pilot study identifying pathways, which are of great importance for providing further hypothesis and direct upcoming research.
Conclusions
We could identify deregulated signaling pathways as a consequence of a directed cellular response to platinum-induced (oxidative) cell stress. In this context, mitogen signal associated pathways, such as MAPK, relaxin, TNF, or Wnt signaling, and energy metabolism regulation via AMPK and mTOR, as well as the mediation of tumor immunogenicity via cytokine release, play important roles. Our results are paving the ground for a better understanding of cellular response and escape mechanisms, carrying a high potential for improved clinical management of patients with MPM.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://dx.doi.org/10.21037/tlcr-21-201
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tlcr-21-201
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tlcr-21-201). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This retrospective study conforms to the principles outlined in the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committee of the Medical Faculty of the University Duisburg-Essen (identifier: 14-5775-BO) and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Faig J, Howard S, Levine EA, et al. Changing pattern in malignant mesothelioma survival. Transl Oncol 2015;8:35-9. [Crossref] [PubMed]
- Gemba K, Fujimoto N, Aoe K, et al. Treatment and survival analyses of malignant mesothelioma in Japan. Acta Oncol 2013;52:803-8. [Crossref] [PubMed]
- Tsao AS, Lindwasser OW, Adjei AA, et al. Current and Future Management of Malignant Mesothelioma: A Consensus Report from the National Cancer Institute Thoracic Malignancy Steering Committee, International Association for the Study of Lung Cancer, and Mesothelioma Applied Research Foundation. J Thorac Oncol 2018;13:1655-67. [Crossref] [PubMed]
- Bueno R, Opitz I. Surgery in Malignant Pleural Mesothelioma. J Thorac Oncol 2018;13:1638-54. [Crossref] [PubMed]
- Baas P, Scherpereel A, Nowak AK, et al. First-line nivolumab plus ipilimumab in unresectable malignant pleural mesothelioma (CheckMate 743): a multicentre, randomised, open-label, phase 3 trial. Lancet 2021;397:375-86. [Crossref] [PubMed]
- Cantini L, Hassan R, Sterman DH, et al. Emerging Treatments for Malignant Pleural Mesothelioma: Where Are We Heading? Front Oncol 2020;10:343. [Crossref] [PubMed]
- Tomek S, Emri S, Krejcy K, et al. Chemotherapy for malignant pleural mesothelioma: past results and recent developments. Br J Cancer 2003;88:167-74. [Crossref] [PubMed]
- Tomek S, Manegold C. Chemotherapy for malignant pleural mesothelioma: past results and recent developments. Lung Cancer 2004;45:S103-19. [Crossref] [PubMed]
- Zucali PA, Giovannetti E, Destro A, et al. Thymidylate synthase and excision repair cross-complementing group-1 as predictors of responsiveness in mesothelioma patients treated with pemetrexed/carboplatin. Clin Cancer Res 2011;17:2581-90. [Crossref] [PubMed]
- Rosell R, Taron M, Barnadas A, et al. Nucleotide excision repair pathways involved in Cisplatin resistance in non-small-cell lung cancer. Cancer Control 2003;10:297-305. [Crossref] [PubMed]
- Olaussen KA, Mountzios G, Soria JC. ERCC1 as a risk stratifier in platinum-based chemotherapy for nonsmall-cell lung cancer. Curr Opin Pulm Med 2007;13:284-9. [Crossref] [PubMed]
- Booton R, Ward T, Ashcroft L, et al. ERCC1 mRNA expression is not associated with response and survival after platinum-based chemotherapy regimens in advanced non-small cell lung cancer. J Thorac Oncol 2007;2:902-6. [Crossref] [PubMed]
- Friboulet L, Barrios-Gonzales D, Commo F, et al. Molecular Characteristics of ERCC1-Negative versus ERCC1-Positive Tumors in Resected NSCLC. Clin Cancer Res 2011;17:5562-72. [Crossref] [PubMed]
- Fujii T, Toyooka S, Ichimura K, et al. ERCC1 protein expression predicts the response of cisplatin-based neoadjuvant chemotherapy in non-small-cell lung cancer. Lung Cancer 2008;59:377-84. [Crossref] [PubMed]
- Hubner RA, Riley RD, Billingham LJ, et al. Excision repair cross-complementation group 1 (ERCC1) status and lung cancer outcomes: a meta-analysis of published studies and recommendations. PLoS One 2011;6:e25164 [Crossref] [PubMed]
- Zimling ZG, Sorensen JB, Gerds TA, et al. Low ERCC1 expression in malignant pleural mesotheliomas treated with cisplatin and vinorelbine predicts prolonged progression-free survival. J Thorac Oncol 2012;7:249-56. [Crossref] [PubMed]
- Bhagwat NR, Roginskaya VY, Acquafondata MB, et al. Immunodetection of DNA repair endonuclease ERCC1-XPF in human tissue. Cancer Res 2009;69:6831-8. [Crossref] [PubMed]
- Kerr KM. Personalized medicine for lung cancer: new challenges for pathology. Histopathology 2012;60:531-46. [Crossref] [PubMed]
- Kamal NS, Soria JC, Mendiboure J, et al. MutS homologue 2 and the long-term benefit of adjuvant chemotherapy in lung cancer. Clin Cancer Res 2010;16:1206-15. [Crossref] [PubMed]
- Jung Y, Lippard SJ. Direct cellular responses to platinum-induced DNA damage. Chem Rev 2007;107:1387-407. [Crossref] [PubMed]
- Koch M, Krieger ML, Stolting D, et al. Overcoming chemotherapy resistance of ovarian cancer cells by liposomal cisplatin: molecular mechanisms unveiled by gene expression profiling. Biochem Pharmacol 2013;85:1077-90. [Crossref] [PubMed]
- Kelly RJ, Sharon E, Hassan R. Chemotherapy and targeted therapies for unresectable malignant mesothelioma. Lung Cancer 2011;73:256-63. [Crossref] [PubMed]
- Walter RF, Vollbrecht C, Werner R, et al. Screening of Pleural Mesotheliomas for DNA-damage Repair Players by Digital Gene Expression Analysis Can Enhance Clinical Management of Patients Receiving Platin-Based Chemotherapy. J Cancer 2016;7:1915-25. [Crossref] [PubMed]
- Ting S, Mairinger FD, Hager T, et al. ERCC1, MLH1, MSH2, MSH6, and betaIII-tubulin: resistance proteins associated with response and outcome to platinum-based chemotherapy in malignant pleural mesothelioma. Clin Lung Cancer 2013;14:558-67 e3. [Crossref] [PubMed]
- Mairinger FD, Werner R, Flom E, et al. miRNA regulation is important for DNA damage repair and recognition in malignant pleural mesothelioma. Virchows Arch 2017;470:627-37. [Crossref] [PubMed]
- Travis WD, Brambilla E, Burke A, et al. WHO classification of tumours of the lung, pleura, thymus and heart. 4th ed. World Health Organization classification of tumours. Lyon: IARC Press, 2015.
- Brierley JD, Gospodarowicz MK, Wittekind C, et al. TNM Classification of Malignant Tumours. 8th ed. Oxford: Wiley, 2017.
- Byrne MJ, Nowak AK. Modified RECIST criteria for assessment of response in malignant pleural mesothelioma. Ann Oncol 2004;15:257-60. [Crossref] [PubMed]
- Ceresoli GL, Chiti A, Zucali PA, et al. Assessment of tumor response in malignant pleural mesothelioma. Cancer Treat Rev 2007;33:533-41. [Crossref] [PubMed]
- Waggott D, Chu K, Yin S, et al. NanoStringNorm: an extensible R package for the pre-processing of NanoString mRNA and miRNA data. Bioinformatics 2012;28:1546-8. [Crossref] [PubMed]
- Available online: www.r-project.org. Accessed 15.12. 2020.
- Available online: http://www.webgestalt.org/. Accessed 15.12. 2020.
- Liao Y, Wang J, Jaehnig EJ, et al. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res 2019;47:W199-W205. [Crossref] [PubMed]
- Thakur B, Ray P. Cisplatin triggers cancer stem cell enrichment in platinum-resistant cells through NF-kappaB-TNFalpha-PIK3CA loop. J Exp Clin Cancer Res 2017;36:164. [Crossref] [PubMed]
- Galluzzi L, Senovilla L, Vitale I, et al. Molecular mechanisms of cisplatin resistance. Oncogene 2012;31:1869-83. [Crossref] [PubMed]
- Shen DW, Pouliot LM, Hall MD, et al. Cisplatin resistance: a cellular self-defense mechanism resulting from multiple epigenetic and genetic changes. Pharmacol Rev 2012;64:706-21. [Crossref] [PubMed]
- Mairinger F, Bankfalvi A, Schmid KW, et al. Digital Immune-Related Gene Expression Signatures In High-Grade Serous Ovarian Carcinoma: Developing Prediction Models For Platinum Response. Cancer Manag Res 2019;11:9571-83. [Crossref] [PubMed]
- Ohmichi M, Hayakawa J, Tasaka K, et al. Mechanisms of platinum drug resistance. Trends Pharmacol Sci 2005;26:113-6. [Crossref] [PubMed]
- Hernandez Losa J, Parada Cobo C, Guinea Viniegra J, et al. Role of the p38 MAPK pathway in cisplatin-based therapy. Oncogene 2003;22:3998-4006. [Crossref] [PubMed]
- Munoz-Galvan S, Felipe-Abrio B, Garcia-Carrasco M, et al. New markers for human ovarian cancer that link platinum resistance to the cancer stem cell phenotype and define new therapeutic combinations and diagnostic tools. J Exp Clin Cancer Res 2019;38:234. [Crossref] [PubMed]
- Kumar S, Kumar A, Shah PP, et al. MicroRNA signature of cis-platin resistant vs. cis-platin sensitive ovarian cancer cell lines. J Ovarian Res 2011;4:17. [Crossref] [PubMed]
- Jin L, Chun J, Pan C, et al. MAST1 Drives Cisplatin Resistance in Human Cancers by Rewiring cRaf-Independent MEK Activation. Cancer Cell 2018;34:315-30.e7. [Crossref] [PubMed]
- Candido J, Hagemann T. Cancer-related inflammation. J Clin Immunol 2013;33:S79-84. [Crossref] [PubMed]
- Fabre JAS, Giustinniani J, Garbar C, et al. The Interleukin-17 Family of Cytokines in Breast Cancer. Int J Mol Sci 2018;19:3880. [Crossref] [PubMed]
- Huber M, Heink S, Pagenstecher A, et al. IL-17A secretion by CD8+ T cells supports Th17-mediated autoimmune encephalomyelitis. J Clin Invest 2013;123:247-60. [Crossref] [PubMed]
- Chen J, Ye X, Pitmon E, et al. IL-17 inhibits CXCL9/10-mediated recruitment of CD8(+) cytotoxic T cells and regulatory T cells to colorectal tumors. J Immunother Cancer 2019;7:324. [Crossref] [PubMed]
- Maeda M, Chen Y, Lee S, et al. Induction of IL-17 production from human peripheral blood CD4+ cells by asbestos exposure. Int J Oncol 2017;50:2024-32. [Crossref] [PubMed]
- Zebedeo CN, Davis C, Pena C, et al. Erionite induces production of autoantibodies and IL-17 in C57BL/6 mice. Toxicol Appl Pharmacol 2014;275:257-64. [Crossref] [PubMed]
- Wang JT, Li H, Zhang H, et al. Intratumoral IL17-producing cells infiltration correlate with anti-tumor immune contexture and improved response to adjuvant chemotherapy in gastric cancer. Ann Oncol 2019;30:266-73. [Crossref] [PubMed]
- Zheng H, Zhang M, Ma S, et al. Identification of the key genes associated with chemotherapy sensitivity in ovarian cancer patients. Cancer Med 2020;9:5200-9. [Crossref] [PubMed]
- Samuel CS, Royce SG, Hewitson TD, et al. Anti-fibrotic actions of relaxin. Br J Pharmacol 2017;174:962-76. [Crossref] [PubMed]
- Ng HH, Shen M, Samuel CS, et al. Relaxin and extracellular matrix remodeling: Mechanisms and signaling pathways. Mol Cell Endocrinol 2019;487:59-65. [Crossref] [PubMed]
- Nair VB, Samuel CS, Separovic F, et al. Human relaxin-2: historical perspectives and role in cancer biology. Amino Acids 2012;43:1131-40. [Crossref] [PubMed]
- Neschadim A, Summerlee AJ, Silvertown JD. Targeting the relaxin hormonal pathway in prostate cancer. Int J Cancer 2015;137:2287-95. [Crossref] [PubMed]
- Yang H, Bocchetta M, Kroczynska B, et al. TNF-alpha inhibits asbestos-induced cytotoxicity via a NF-kappaB-dependent pathway, a possible mechanism for asbestos-induced oncogenesis. Proc Natl Acad Sci U S A 2006;103:10397-402. [Crossref] [PubMed]
- Carbone M, Bedrossian CW. The pathogenesis of mesothelioma. Semin Diagn Pathol 2006;23:56-60. [Crossref] [PubMed]
- Liu J, Sun RW, Leung CH, et al. Inhibition of TNF-alpha stimulated nuclear factor-kappa B (NF-kappaB) activation by cyclometalated platinum(II) complexes. Chem Commun (Camb) 2012;48:230-2. [Crossref] [PubMed]
- Inoki K, Kim J, Guan KL. AMPK and mTOR in cellular energy homeostasis and drug targets. Annu Rev Pharmacol Toxicol 2012;52:381-400. [Crossref] [PubMed]
- Carling D. AMPK signalling in health and disease. Curr Opin Cell Biol 2017;45:31-7. [Crossref] [PubMed]
- Wang Z, Wang N, Liu P, et al. AMPK and Cancer. Exp Suppl 2016;107:203-26. [PubMed]
- Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer 2009;9:563-75. [Crossref] [PubMed]
- Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell 2007;12:9-22. [Crossref] [PubMed]
- Mayer IA, Arteaga CL. The PI3K/AKT Pathway as a Target for Cancer Treatment. Annu Rev Med 2016;67:11-28. [Crossref] [PubMed]
- Wilson SM, Barbone D, Yang TM, et al. mTOR mediates survival signals in malignant mesothelioma grown as tumor fragment spheroids. Am J Respir Cell Mol Biol 2008;39:576-83. [Crossref] [PubMed]
- Mochizuki D, Adams A, Warner KA, et al. Anti-tumor effect of inhibition of IL-6 signaling in mucoepidermoid carcinoma. Oncotarget 2015;6:22822-35. [Crossref] [PubMed]
- Nor C, Zhang Z, Warner KA, et al. Cisplatin induces Bmi-1 and enhances the stem cell fraction in head and neck cancer. Neoplasia 2014;16:137-46. [Crossref] [PubMed]
- Granville CA, Memmott RM, Gills JJ, et al. Handicapping the race to develop inhibitors of the phosphoinositide 3-kinase/Akt/mammalian target of rapamycin pathway. Clin Cancer Res 2006;12:679-89. [Crossref] [PubMed]
- Salminen A, Kauppinen A, Kaarniranta K. AMPK activation inhibits the functions of myeloid-derived suppressor cells (MDSC): impact on cancer and aging. J Mol Med (Berl) 2019;97:1049-64. [Crossref] [PubMed]
- Uehara T, Eikawa S, Nishida M, et al. Metformin induces CD11b+-cell-mediated growth inhibition of an osteosarcoma: implications for metabolic reprogramming of myeloid cells and anti-tumor effects. Int Immunol 2019;31:187-98. [Crossref] [PubMed]
- Qin G, Lian J, Huang L, et al. Metformin blocks myeloid-derived suppressor cell accumulation through AMPK-DACH1-CXCL1 axis. Oncoimmunology 2018;7:e1442167 [Crossref] [PubMed]
- Trikha P, Plews RL, Stiff A, et al. Targeting myeloid-derived suppressor cells using a novel adenosine monophosphate-activated protein kinase (AMPK) activator. Oncoimmunology 2016;5:e1214787 [Crossref] [PubMed]
- Mi L, Zhou Y, Wu D, et al. ACSS2/AMPK/PCNA pathwaydriven proliferation and chemoresistance of esophageal squamous carcinoma cells under nutrient stress. Mol Med Rep 2019;20:5286-96. [PubMed]
- Dauer P, Nomura A, Saluja A, et al. Microenvironment in determining chemoresistance in pancreatic cancer: Neighborhood matters. Pancreatology 2017;17:7-12. [Crossref] [PubMed]
- Chou CW, Wang CC, Wu CP, et al. Tumor cycling hypoxia induces chemoresistance in glioblastoma multiforme by upregulating the expression and function of ABCB1. Neuro Oncol 2012;14:1227-38. [Crossref] [PubMed]
- Cruz-Bermudez A, Laza-Briviesca R, Vicente-Blanco RJ, et al. Cancer-associated fibroblasts modify lung cancer metabolism involving ROS and TGF-beta signaling. Free Radic Biol Med 2019;130:163-73. [Crossref] [PubMed]
- Zhang W, Bouchard G, Yu A, et al. GFPT2-Expressing Cancer-Associated Fibroblasts Mediate Metabolic Reprogramming in Human Lung Adenocarcinoma. Cancer Res 2018;78:3445-57. [Crossref] [PubMed]
- Alexander J, Cukierman E. Stromal dynamic reciprocity in cancer: intricacies of fibroblastic-ECM interactions. Curr Opin Cell Biol 2016;42:80-93. [Crossref] [PubMed]
- Heneberg P. Paracrine tumor signaling induces transdifferentiation of surrounding fibroblasts. Crit Rev Oncol Hematol 2016;97:303-11. [Crossref] [PubMed]
- Spaeth EL, Dembinski JL, Sasser AK, et al. Mesenchymal stem cell transition to tumor-associated fibroblasts contributes to fibrovascular network expansion and tumor progression. PLoS One 2009;4:e4992 [Crossref] [PubMed]
- Xu X, Cheng L, Fan Y, et al. Tumor Microenvironment-Associated Immune-Related Genes for the Prognosis of Malignant Pleural Mesothelioma. Front Oncol 2020;10:544789 [Crossref] [PubMed]
- Rapp J, Jaromi L, Kvell K, et al. WNT signaling - lung cancer is no exception. Respir Res 2017;18:167. [Crossref] [PubMed]
- Shapiro M, Akiri G, Chin C, et al. Wnt pathway activation predicts increased risk of tumor recurrence in patients with stage I non-small cell lung cancer. Ann Surg 2013;257:548-54. [Crossref] [PubMed]
- Stewart DJ. Wnt signaling pathway in non-small cell lung cancer. J Natl Cancer Inst 2014;106:djt356 [Crossref] [PubMed]
- Gattinoni L, Zhong XS, Palmer DC, et al. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat Med 2009;15:808-13. [Crossref] [PubMed]
- Reuter S, Beckert H, Taube C. Take the Wnt out of the inflammatory sails: modulatory effects of Wnt in airway diseases. Lab Invest 2016;96:177-85. [Crossref] [PubMed]
- Staal FJ, Luis TC, Tiemessen MM. WNT signalling in the immune system: WNT is spreading its wings. Nat Rev Immunol 2008;8:581-93. [Crossref] [PubMed]
- Li X, Xiang Y, Li F, et al. WNT/beta-Catenin Signaling Pathway Regulating T Cell-Inflammation in the Tumor Microenvironment. Front Immunol 2019;10:2293. [Crossref] [PubMed]
- Holtzhausen A, Zhao F, Evans KS, et al. Melanoma-Derived Wnt5a Promotes Local Dendritic-Cell Expression of IDO and Immunotolerance: Opportunities for Pharmacologic Enhancement of Immunotherapy. Cancer Immunol Res 2015;3:1082-95. [Crossref] [PubMed]
- Li J, He X, Zou Y, et al. Mitochondria-targeted platinum(ii) complexes: dual inhibitory activities on tumor cell proliferation and migration/invasion via intracellular trafficking of beta-catenin. Metallomics 2017;9:726-33. [Crossref] [PubMed]



