Real-time autOmatically updated data warehOuse in healThcare (ROOT): an innovative and automated data collection system
Introduction
Real-world data (RWD) and real-world evidence (RWE) play an increasing important role in healthcare decisions. For example, they are used by the US Food and Drug Administration to monitor post-marketing safety and adverse events and to support regulatory decisions. Additionally, the healthcare community uses this type of data to support coverage decisions; and develop clinical practice guidelines and decision-making support tools. Developers of medical products also use RWD and RWE to support clinical trial designs and observational studies to generate innovative treatment approaches (1). However, several barriers that prevent the integration of medical data in the real world, including the complexity caused using different methods of recordkeeping, formats, reference values used, and privacy issues as well as missing values. Therefore, updating a database can be a time-consuming challenge (2).
Recently, the American Society of Clinical Oncology (ASCO) launched the minimal common oncology data elements (mCODE) project to facilitate cancer data interoperability and to improve the overall quality of cancer data for patient care and research (3). Data need to be de-identified. This project will provide a common data language and an open-source, nonproprietary data model based on Fast Healthcare Interoperability Resources for interconnectivity across electronic medical record (EMR) systems. For this project (mCODE), standardized data were collected computably for integration with data from other patients and then analyzed for best practices. Data collection needs to be streamlined so that it does not burden busy clinicians. Maintaining the security and privacy of patients is also essential. The mCODE project was discussed and launched in earnest at the ASCO’s annual meeting in 2019. ASCO is currently discussing the definitions of data elements and practical implementation methods.
The Clinical Data Warehouse (CDW) is a real-time database that aggregate data from various clinical sources to provide the entirety of clinical data for each patient from a unified view (4). CDW, which was initially developed for clinical research and hospital financial analyses, is now evolving to support efforts to improve the quality of healthcare services. For example, CDW provides information on activity trends and the evolution of case mixes. However, the clinical data extracted from CDW contain structured data and natural language generated during daily clinical practice. Therefore, applying CDW data to clinical research is challenging because they are not always structured and formatted to facilitate the retrieval of important information (2,5-8).
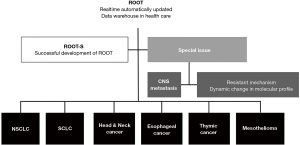
An innovative data collection system with a specialized algorithm, called Real-time autOmatically updated data warehOuse for healThcare (ROOT), has been developed to comprehensively and systematically collect CDW data from cancer cohorts.
Methods
ROOT was developed to automatically extract and update CDW data in real-time. ROOT contain comprehensive clinical information from patients with six histologically confirmed types of solid tumors—head and neck cancer, esophageal cancer, non-small cell lung cancer (NSCLC), small cell lung cancer (SCLC), thymic carcinoma, and mesothelioma—diagnosed at the Samsung Medical Center (SMC) from January 2008 to January 2020. It includes results from routine laboratory tests, radiological examinations, histopathological examinations, and clinical features. It also includes particular issues of interest, e.g., central nervous system CNS metastases and resistance mechanisms to targeted therapies (Figure 1). Histopathological and molecular/genomic data from longitudinal biopsies have also been collected and integrated for the resistance mechanisms. For each cancer cohort, key elements within six main areas were defined, collecting more than 700 kinds of clinical data to show the journey of each cancer patient (Table 1).
Table 1
| Elements | Areas |
|---|---|
| Patients | Age*; sex*; performance status*; smoking history*; family history#; co-morbidity (HTN, DM, hepatitis, Tbc, cardiovascular disease, cerebral disease, thyroid disorder)# |
| Disease | Histology*; TNM stage (c, yp, p)*; location of primary tumor#; metastatic site (brain*, bone&, lung&, liver&, pleura&, leptomeningeal seeding&) |
| Genomics* | EGFR mutation [PCR clamping: tissue and liquid (blood, body fluid)]; ALK rearrangement (IHC, FISH); PD-L1 IHC; ROS1 (RT-PCR, IHC); BRAF (RT-PCR); KRAS (IHC); TRK (IHC) NGS (targeted sequencing; CANCER Scan, Perseq Oncomine); P16; repeated biopsies |
| Labs# | Tumor marker; CBC; chemistry; electrolyte; LD; CRP |
| Treatment* | Surgery: aim: curative vs. palliative; types of surgery (lobectomy, pneumonectomy): VATS or open, Craniotomy, VP shunt, Omaya insertion |
| Radiotherapy: aim: curative vs. palliative; location: primary cancer site/metastasis site/brain | |
| Stereotactic radiosurgery | |
| Chemotherapy: adjuvant/neoadjuvant/definitive/palliative/salvage treatment | |
| Clinical trial | |
| Outcomes | RFS*; PFS*; TTNT*; OS*; RR*; side effects (clinical symptoms, labs, radiation pneumonitis, drug-induced pneumonitis)& |
*: level I; &: level II; #: level III. HTN, hypertension; DM, diabetes; Tbc, tuberculosis; EGFR, epidermal growth factor receptor; ALK, anaplastic lymphoma kinase; IHC, immunohistochemistry; FISH, fluorescent in situ hybridization; PD-L1, programmed death-ligand 1; NGS, next-generation sequencing; VATS, video-assisted thoracoscopy; RFS, relapse-free survival; PFS, progression-free survival; TTNT, time to next treatment; OS, overall survival; RR, response rate.
Development of the algorithm
The development of ROOT took place in three steps: program development, validation, and updating.
Program development
A program was developed for data extraction, transformation, and loading (ETL) using a standard query language (SQL) and the SAP Data Services Designer 14.2.1.224 (a developer tool used to create objects through data mapping, transformation, and logic). The primary sources of data were extracted from the EMR system used at SMC (Darwin-Med). ETL is a general procedure for copying data from one or more sources into a destination system that presents the data differently. Data extraction involves extracting data from homogeneous or heterogeneous sources; data transformation involves cleaning the data and transforming them into a storage format appropriate for querying and analysis; and data loading is the insertion of the transformed data into the final target database, such as an operational data repository, a data mart, or a data warehouse.
Data source and data flow
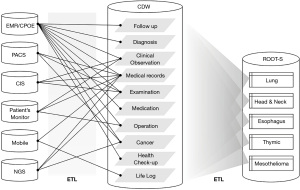
The database management system (DBMS) included Darwin-MED (version 11.6), which was an EMR system, and Darwin-C (version1.0.7404), which was a CDW system at SMC. Figure 2 indicates a data flowchart using various references and sources for ROOT. CDW is organized by categories and data items, usually as formatted information. ROOT is designed for each of the six types of cancer, using data stored and integrated from various CDW data sources. Thus, the list of variables for the six types of cancer in ROOT all differs from one another. ROOT included all new patients diagnosed with histologically confirmed six-type of cancer in daily clinic every 24 h. ROOT used the diagnostic code of the International Classification of Disease (ICD) and Korean standard classification of disease (KCD), the national cancer registry, histologic data, and the clinician’s decision recorded in EMR for the final diagnosis of primary cancer. If new patients with NSCLC visited the clinic, they were updated automatically on ROOT when receiving any test or recording in the EMR system. In the system of CDW, patients were de-identified to protect their privacy.
Algorithm development
The algorithm to extract necessary data and define the priority of data from various sources and references were developed. For examples of natural language processing, consider data from metastatic sites during radiological examination (e.g., MRI, CT), which includes natural language such as ‘probable, possible, and likely new metastatic sites’. The definitions of the subgroup items within the six main regions were determined, and sample data were extracted to check whether the items met the criteria. The process of extracting and reviewing the sample data was repeated several times during the program development. When an item appears in multiple sources, the priority of each source was determined, and changes in importance were tested by reviewing sample data according to the designed criteria. The source and reference of each variable are described in ROOT. Figure S1 indicates an example of the Data Services Designer (version 14.2), and Figure S2 indicate an instance of the algorithm used for data extraction.
Data extraction
ROOT extracted data from CDW-derived Darwin-Med (original data source) according to the specialized algorithm. ROOT conducted daily extraction using programmed SQL and SAP Data Services Designer 14, a developer tool used to create objects through data mapping, transformation, and logic.
Transform
ROOT systemically transformed the different types of data, including free text and numerical data, into a storage format and structure appropriate for querying and analysis systematically.
Load
ROOT data extracted using the program were automatically uploaded to the final data destination every 24 h.
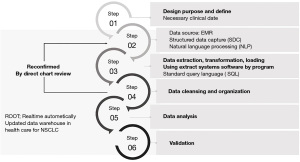
Figure 3 briefly indicates how we developed ROOT and conducted data quality management (DQM) through six steps. Step one was the cohort design. Step two confirmed whether the data source included structured data capture or natural language processing; the algorithm was developed using SQL. Step three included data extraction, transformation, and loading. Step four comprised data cleansing and organization processes. Step five included data analysis, and Step six involved validation. From Steps two through four, the reliability of the data was confirmed through a direct chart review. During the feedback step, the algorithm was modified several times to develop the best version.
Validation process
First, to automatically validate that the algorithm works, the validation sets of patients diagnosed and treated in 2018 were designated. We established how many data points were extracted by developing an algorithm to run between the test set and the validation set. Second, 700 variables were defined according to their degree of importance: Level I (most important), Level II (important), and Level III (less important). To confirm the validity of the data extracted using the algorithm, external analytics designed a random table for randomized patients. Using a direct chart review of the CDW screen (without exposure of personal data such as patient ID), the data’s accuracy was confirmed by comparing the extracted data with the real data of randomly selected patients. The critical elements in Level I were verified using direct chart review to ensure that they were more than 95% accurate, Level II data required to be more than 90% accurate, and Level III data required to be more than 80% accurate. The validation process was repeated several times during program development and this validation continued as the algorithm was updated.
Update process
Planning for ROOT began in August 2018, and the first concept was finalized in February 2019. In February 2020, the validation process of the developed algorithm was re-confirmed and updated. To continue to validate and update ROOT periodically was planned.
Ethics statement
This study was reviewed and approved by the Institutional Review Board (IRB) at SMC (IRB No. 2018-05-130). The trial was performed according to the Helsinki Declaration (as revised in 2013). This study is waived from informed consent due to de-identification.
Statistical analysis
The cutoff date for the data to be included in the analysis was January 2020. ROOT is designed for automatic calculation and updating of cancer-specific outcomes. The response rate, side effects, relapse-free survival (RFS), progression-free survival (PFS), and overall survival (OS) data were automatically calculated and updated every 24 h. RFS is calculated from the first day of curative-intent treatment, such as surgery or definitive radiotherapy, to either the date of relapse, the last follow-up date (LFD), or death resulting from any cause. PFS was computed from the first treatment date to either the date of progression, LFD, or death from any cause. The time to next treatment is calculated from the first date of treatment to the date of next treatment. OS is defined as the time from the date of diagnosis to either LFD or death from any cause.
Results
Clinical data were collected from 67,617 patients (28,954 with NSCLC, 2,540 with SCLC, 30,035 with head and neck cancer (23,578 with thyroid cancer), 4,950 with esophageal cancer, 966 with thymic cancer, and 172 with mesothelioma) diagnosed at SMC from January 2008 to January 2020.
Composition of cohort: six main areas, key elements, and clinical data
The detailed items within the six main areas of patients’ cancer journeys—patient, disease, genomics, laboratory results, treatment, and outcomes—are listed in Table 1. The key elements in the six main areas, evaluated the quality of the raw data extracted from different sources, and then organized the data by checking their priority. Within these six primary areas, more than 70 essential elements and 700 clinical data points were obtained. The key elements comprise of clinical data with clinical significance.
Final key panel and comprehensive panel
A final key panel and a comprehensive panel for each cancer cohort was constructed using the key elements and variables in the six main areas. The final key panel is likely to be the most clinically useful, in which different vital elements are organized by type. Comprehensive panels can be combined as desired by the user. The comprehensive panels were organized separately so that users can easily compile raw data according to their requirements and research purposes. For example, if data on a patient’s performance status were extracted from the EMR, before extraction, the raw data could be in the form of fill-in-the-blank responses or free text. Additionally, performance status could be extracted at different time points, depending on the patient’s treatment. For example, because the comprehensive panel contains all information on all types of chemotherapy, it is possible to obtain clinical data for patients using specific regimens in specific settings. The data were classified as clearly defined data (can be used as automatically extracted), structured data (need clarification or modification), and unstructured data (usually need natural language processing) (Table 2). The comprehensive clinical data were integrated to minimize and compensate for the effect of missing values.
Table 2
| Clearly defined dataa | Structured datab | Unstructured datac |
|---|---|---|
| Age | ECOG (EMR: FB) | ECOG (EMR: text) |
| Sex | Smoking (EMR: FB) | Smoking (EMR: text) |
| Dates (birthday, date of first diagnosis, date of first treatment, surgery, death) | Family history (EMR: FB) | Family history (EMR: text) |
| Co-morbidity (diagnostic codes) | Co-morbidity (EMR: FB) | Co-morbidity (EMR: text or medication) |
| Blood test | ||
| Pathology result | Pathology result (out-of-hospital) | |
| Mutation test (EGFR/ALK/PD-L1 in-house setting) | Mutation test (NGS/clinical trial, other hospital) | |
| TNM staging (EMR: FB) | TNM staging (image, EMR: text) | |
| Type of surgery/radiation dosage, fraction, location/chemotherapy regimen | Image result (brain metastasis, LMS, metastasis site) |
a, data that can be used as they are automatically extracted from the EMR; b, results that are clear but need clarification and modification; c, data used to synthesize the results extracted by various methods or natural language processing. FB, fill in the blank.
Level I key elements
The key elements were classified as Level I (most important), Level II (important), and Level III (less important) (Table 1). The vital elements under the main area “patients,” a patient’s age, sex, performance status, and smoking history, were classified as Level I. For performance status and smoking history, the data were extracted from the outpatient records, hospitalization records, surgical records, and nursing records. Areas of disease, histology, TNM (clinical, post-neoadjuvant, pathological stage), and brain metastasis constitute Level I information. For the key elements of brain metastasis, data on the treatments for brain metastasis, such as stereotactic radiosurgery (SRS), whole-brain radiotherapy (WBRT), craniotomy, tumor removal, intrathecal chemotherapy, and ventriculoperitoneal shunt operations, were collected. Additionally, natural language data were extracted from the EMR and test results from brain MRI. For brain metastasis, cytology and molecular study of CSF (esp. EGFR mutation) were included. The data of brain metastasis from radiology reports of CT or MRI. ROOT performed parsing pre-defined terminology or sentence such as “multiple brain metastasis existed in the cerebellum and frontal lobe”. However, false-positives, such as a sentence of “no evidence of brain metastasis”, the existence of brain metastasis was differentiated using various operational definitions. All genomic data were categorized as Level I. When collecting data about the significant driver mutations, such as EGFR, ALK, ROS-1, PD-L1, and BRAF, all sources of data were extracted from the EMR. Test results were collected from the institute using data capture and other hospitals’ results by the natural language processing of EMR. For example, PNAclampTM Kits and real-time polymerase chain reactions, COBAS, and next-generation sequencing (NGS) of tissue or liquid were collected for detection of EGFR mutations. Outsourced results were included from referral hospitals using the natural language processing and capture method for EMR records. All treatments and outcomes are designated as Level I. We collected information about all kinds of chemotherapy regimens and clinical trials conducted at SMX, including sponsor-initiated trials and investigator-initiated trials. The NSCLC cohort contains data about more than 60 chemotherapy regimens and 150 clinical trials. All diagnostic tests and treatments were checked against prescriptions, and it was verified that they had actually been conducted. The prescription codes (diagnostic codes, test codes, codes for chemotherapy, surgery, radiotherapy, and other treatments) are updated periodically.
Survival data include information from the national cancer registry, national health insurance, and our own institution. The survival data in the national cancer register were updated every year. However, data regarding the suspension of qualification for national health insurance are updated every 24 h, and the survival data of the institution were updated in real-time. By using survival data from different sources, we can update information about patients who did not die at the hospital. The algorithm selects the most reliable and recently updated information from the three survival information sources (national cancer register, national health data, and our own institute). PFS and DFS data were collected using data of EMR and radiographic tests. ROOT captured the data of progression in EMR, which the clinician recorded disease progression and switching chemotherapy regimen. ROOT also captured data of radiological progression. In case of switching chemotherapy regimens due to adverse events, ROOT captured why chemotherapy was switched. The data of objective response of each chemotherapy was extracted from EMR through the clinician’s judgment and radiological assessment.
Result of the validation process
We randomly selected 200 patients from each cohort to validate ROOT. An independent reviewer extracted all clinical data in the actual EMR. For example, the clinical stage (Level I) was confirmed in 95.6% of NSCLC patients. The history of chemotherapy was confirmed in all patients, except for few of blinded clinical trial registrants. Survival data indicated more than 95% accuracy and had no missing values. For other examples of validation of ROOT, the data on recurrence included local and distant recurrence after curative-intent surgery in a patient with stage I–III NSCLC. By direct chart review, how ROOT extracted data of recurrence from multiple sources, including EMR and radiologic examination such as CT or MRI. The accuracy of data of recurrence of ROOT was 95% compared to direct chart review by the medical oncologist. In patients diagnosed with early-stage NSCLC and received curative-intent surgery, the data of DFS and site of recurrence in ROOT and direct-chart review by medical oncologist were compared.
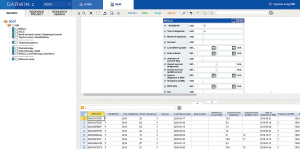
User-friendly cohort system
Figure 4 indicates the actual ROOT interface in the CDW program. ROOT provides a final key panel containing the Level I key elements from the six main areas and a comprehensive panel in which more than 700 clinical data items were structured and organized. Researchers can easily extract data suitable for their research purposes from the comprehensive panel. As indicated in Figure 4, the user first clicks on the needed clinical data and then enters the required search conditions. For example, they might select a required clinical setting or a group that has undergone homogeneous treatment, and the algorithm will comprehensively extract those clinical data. De-identified search results can be downloaded in an Excel format and are updated every 24 h. Additionally, clinical outcomes, such as RFS, PFS, time to subsequent treatment, and OS, are automatically calculated every 24 h.
Discussion
ROOT is a unique algorithm developed to capture comprehensive information in a systematic, organized, and timely manner and to automatically update a self-renewing cohort over time. ROOT updated all cancer-specific data, including test results and outcomes.
ROOT uses CDW for data collection and organization. Several studies have used CDW as a cohort for clinical disease information (6,9,10). CDW already includes large medical data from various sources in the EMR. However, CDW is not particularly useful on its own, because it provides only a simple list that needs to be manually analyzed. Previous studies have demonstrated clinical outcomes using CDW, but most of these studies only analyzed simple variables. It is challenging to obtain and analyze large medical data, especially for cancer patients, for the following reasons. The usual pathways for cancer treatment are complicated and include diagnostics, new drugs/drug combinations, new subtypes of molecularly defined entities, and new techniques of radiotherapy and surgery. Moreover, in the real world, individual patients rarely follow a standard path. The complexity of modern treatment options can make optimal treatment challenging to sustain. Unexpected situations can occur in each patient before, during, or after treatment. Furthermore, reimbursement issues, including restrictions imposed by payers or providers (facilities/guidelines), can complicate matters.
To the best of our knowledge, we are the first to report the successful development of a disease-specific algorithm, and it has many advantages. First, ROOT uses innovative technology to enhance data management. It could enhance healthcare, both internationally and inter-institutionally, by elevating medical care and research quality. It provides data for physicians to support informed clinical decisions and research. Perhaps its most important characteristic is that it could be applied and used in all medical fields. For example, in the case of epidemic outbreaks of emerging diseases (e.g., COVID-19) or new diseases, physicians could use the developed algorithm to obtain, and update clinical information in real-time. Second, the data in ROOT are updated automatically every 24 h, meaning that researchers can always access the most recently updated data, including diagnostic test results and outcomes. This should enable the development of appropriate research designs. Third, the data configuration is user-friendly regarding data collection and analysis. ROOT collects usual clinical practice data without needing any additional time or effort regarding cohort construction or data collection. There were level I key elements that were common in six-type of cancers. The common elements included TNM age, ECOG performance status, TNM staging system, line of palliative chemotherapy, treatment outcome, and genomic profile from NGS. As specified data for each cohort of six-type of cancer, clinical data were obtained and defined as levels I, II, III. For example, cohort of NSCLC, the status of major driver mutations, such as EGFR, ALK, BRAF, and ROS-1 mutation, outcomes of matched targeted therapy, a type of resistance mechanism, and method to detect the resistant mechanism were included in level I key element. In the cohort of head and neck cancer, EBV and HPV status were included. In patients with thymic epithelial tumors, Masaoka stage and WHO classification were included in level I key element. For extraction of TNM staging, to determine the priority of date from a different time and multiple sources is needed. In the case of early-stage NSCLC who received curative-intent surgery, there were at least five data of TNM. It was validated several times for each clinical scenario, which data of TNM stage in EMR represented final decisions. Fourth, the data were entirely de-identified and were free from privacy issues. Lastly, all patients newly diagnosed at SMC with any of the six types of cancer detailed here were automatically registered, and their data are collected every 24 h. The system is therefore sustainable.
While developing ROOT, based on CDW, we were confronted with many challenges, which were addressed through a close collaboration between treating oncologists and data scientists. The first challenge was to collect high-quality data with minimal missing values from various sources. The second challenge, which represented the most challenging part of development, was the complicated cleaning and organizing the extracted data. The extracted data were cleaned and organized for clinical research using case report forms in Excel format. Data files were configured for immediate use regarding data analysis and clinical research. For example, it is hard to format test results, such as NGS, for Excel. Therefore, we planned to use links at an early stage, but that led to problems with the server capacity. A simple summary of results the results of NGS was instead implemented as structured information. This currently constitutes the most updated version of ROOT, but it is an evolving format. Thirdly, the last but least challenging task was the validation process. The data review was repeated manually using randomly selected patients and then amended the corresponding algorithm accordingly. The ROOT data for some cancer types were analyzed (11,12) as part of that validation process. This confirmed that the results are comparable with historical data published elsewhere (manuscript in preparation) (Table S1 and Figure S3).
The ROOT is still currently in the early stages and thus has several constraints in terms of the sensitivity and data specificity. Therefore, development, and improvement of the algorithm is still ongoing. Another constraint of ROOT is that though the algorithm can be applied to other organs or other types of cancer (or other diseases), it is necessary to check whether problems exist when applied to these different scenarios.
Conclusions
This is the first landmark study to report the successful development of an algorithm to retrieve, analyze, and automatically update CDW data about a cohort of cancer patients in real-time. Thus, ROOT can pave the way to precision medicine by facilitating data retrieval/analysis/exchange and developing future research on time. It also effectively conveys essential clinical information and state-of-the-art treatment pathways to clinicians. Thus, large medical data are expected to play an essential role as real-world evidence in the future.
Acknowledgments
Seung-ho Sin and Gun-Hee-Jo developed the program and algorithm as senior data scientists. Hyeonji Ko and Jiyeon Kim performed a direct chart review for validation.
Funding: This study was supported by a grant from the Korean Society of Medical Oncology (KSMO) 2021. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Ministry of Science and ICT (No. NRF-2021R1F1A1054782).
Footnote
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tlcr-21-531
Peer Review File: Available at https://dx.doi.org/10.21037/tlcr-21-531
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tlcr-21-531). HAJ receives a grant from the Korean Society of Medical Oncology (KSMO) 2021, National Research Foundation of Korea (NRF) grant funded by the Ministry of Science and ICT (No. NRF-2021R1F1A1054782), and consulting fees from AMCA. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was reviewed and approved by the Institutional Review Board (IRB) at SMC (IRB No. 2018-05-130). The trial was performed according to the Helsinki Declaration (as revised in 2013). This study is waived from informed consent due to de-identification.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- ElZarrad MK, Corrigan-Curay J. The US Food and Drug Administration's Real-World Evidence Framework: A Commitment for Engagement and Transparency on Real-World Evidence. Clin Pharmacol Ther 2019;106:33-5. [Crossref] [PubMed]
- Obermeyer Z, Emanuel EJ. Predicting the Future - Big Data, Machine Learning, and Clinical Medicine. N Engl J Med 2016;375:1216-9. [Crossref] [PubMed]
- mCODE™: Minimal Common Oncology Data Elements. Available online: https://mcodeinitiative.org/ (accessed 5 June 2020).
- Karami M, Rahimi A, Shahmirzadi AH. Clinical data warehouse: an effective tool to create intelligence in disease management. Health Care Manag (Frederick) 2017;36:380-4. [Crossref] [PubMed]
- Margolis R, Derr L, Dunn M, et al. The National Institutes of Health's Big Data to Knowledge (BD2K) initiative: capitalizing on biomedical big data. J Am Med Inform Assoc 2014;21:957-8. [Crossref] [PubMed]
- Jannot AS, Zapletal E, Avillach P, et al. The Georges Pompidou University Hospital Clinical Data Warehouse: A 8-years follow-up experience. Int J Med Inform 2017;102:21-8. [Crossref] [PubMed]
- Meyer AM, Olshan AF, Green L, et al. Big data for population-based cancer research: the integrated cancer information and surveillance system. N C Med J 2014;75:265-9. [Crossref] [PubMed]
- Meyer AM, Carpenter WR, Abernethy AP, et al. Data for cancer comparative effectiveness research: past, present, and future potential. Cancer 2012;118:5186-97. [Crossref] [PubMed]
- Kim HS, Kim H, Jeong YJ, et al. Development of Clinical Data Mart of HMG-CoA Reductase Inhibitor for Varied Clinical Research. Endocrinol Metab (Seoul) 2017;32:90-8. [Crossref] [PubMed]
- de Mul M, Alons P, van der Velde P, et al. Development of a clinical data warehouse from an intensive care clinical information system. Comput Methods Programs Biomed 2012;105:22-30. [Crossref] [PubMed]
- Jung HA, Hong S, Park J, et al. Successful Development of Realtime Automatically Updated Data Warehouse in Health Care (ROOT-S). J Thorac Oncol 2019;14:S328. [Crossref]
- Jung HA, Sun JM, Lee SH, et al. Ten-year patient journey of stage III non-small cell lung cancer patients: A single-center, observational, retrospective study in Korea (Realtime autOmatically updated data warehOuse in healTh care; UNIVERSE-ROOT study). Lung Cancer 2020;146:112-9. [Crossref] [PubMed]