
Elevated expression of miR-494-3p is associated with resistance to osimertinib in EGFR T790M-positive non-small cell lung cancer
Introduction
Lung cancer causes the highest death toll of all cancers worldwide (1-3). Activating mutations in the gene encoding the epidermal growth factor receptor (EGFR) is one of the most common targetable genetic aberrations in non-small cell lung cancer (NSCLC) (4), occurring in about 15–20% of lung adenocarcinomas in Europe and North America and in up to 40–50% in Asia (5,6). Activating mutations in EGFR occur mainly as either small deletions in exon 19 or point mutations in exon 21 (7). Tumors harboring activating mutations in EGFR are amenable for therapeutic intervention using tyrosine kinase inhibitors (TKIs) (8). First-generation EGFR TKIs include adenosine triphosphate (ATP)-competitive kinase inhibitors erlotinib and gefitinib and second-generation irreversible inhibitors afatinib and dacomitinib, both generations showing clinical benefit in patients with activating mutations in EGFR (9-12). However, patients eventually develop resistance to these inhibitors, which is frequently accompanied by a gatekeeper mutation, T790M, in exon 20 (13-17). This has led to the development of a third-generation EGFR TKI osimertinib, which irreversibly binds to the C797 residue and thereby targets both activating mutations as well as EGFR molecules with the T790M mutation (18,19). Osimertinib has displayed superior clinical outcome to first-generation EGFR TKIs in clinical trials and is currently a standard choice for advanced stage NSCLC patients with mutations in EGFR (20,21).
MicroRNAs (miRNAs) are small non-coding RNAs of approximately 22 nucleotides in length. miRNAs target a plethora of mRNAs by binding to 3'untranslated regions (3'UTRs) causing mRNA degradation or protein translational inhibition (22). Numerous miRNAs have an impact on the progression of cancer, either as tumor promoting or tumor suppressing actors (23). However, very few studies have investigated the functional role of miRNAs in treatment-refractory cancers. Depending on the target repertoire, specific miRNAs may be particularly suited for therapeutic modulation in treatment-refractory cancers (24,25). miRNAs may be modulated in a therapeutic setting either as mimic molecules, in case the endogenous miRNA has tumor suppressive properties, or through miRNA-inhibitors if the endogenous miRNA plays a tumor-promoting role. The latter is commonly achieved through various chemical synthesis approaches (26,27). We recently demonstrated that specific inhibition of miR-100-5p could restore sensitivity to TKIs targeting echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase (EML4-ALK) positive NSCLC cells in vitro (28).
Here, we aimed to investigate the involvement of miRNAs in resistance to EGFR TKIs in vitro and whether specific miRNAs could serve as markers for EGFR TKI resistance in the clinical setting. We present the following article in accordance with the MDAR reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-21-955/rc).
Methods
Cell culture
NSCLC EGFR mutant parental cell lines NCI-H1975 and HCC827 (ATCC: CRL-5908 and CRL-2868, respectively) and osimertinib-refractory cell lines (29) were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum in a 5% CO2-humidified incubator at 37 ℃ and passaged when reaching sub-confluent conditions.
Chemicals
EGFR TKI osimertinib was purchased from Selleckchem (Cat# S7297; Houston, TX, USA).
Cell viability assay
Parental and TKI-resistant NCI-H1975 and HCC827 cells were plated in triplicates, 4,000 cells in each replicate, in 96-well plates in the presence of 1, 10, 100, and 1,000 nM TKI or in dimethyl sulfoxide (DMSO). Cell viability was scored in a luminometer 72 hours post plating through Cell-titer Glo cell viability assay according to manufacturer’s instruction (Promega, Madison, WI, USA, Cat# G7571). For miRNA mimic experiment, parental HCC827 cells were plated in triplicates for each condition, 2,500 cells in each replicate, in 96-well plates and reverse transfected using Lipofectamine RNAiMAX (ThermoFisher, Waltham, MA, USA) with 1 nM mimic of miR-494-3p (MC12409, ThermoFisher) or with 1 nM negative control mimic (AM17111, ThermoFisher). Osimertinib (100 nM) or DMSO was added 24 hours post transfection. Cell viability was scored in a luminometer 72 hours post osimertinib/DMSO treatment through Cell-titer Glo cell viability assay according to manufacturer’s instruction (Promega, Cat# G7571). For miRNA/DNA experiment, parental HCC827 cells were plated in triplicates for each condition, 5,000 cells in each replicate, in 96-well plates and reverse transfected using Lipofectamine 2000 (ThermoFisher) with 1 nM mimic of miR-494-3p (MC12409, ThermoFisher), with 1 nM negative control mimic (AM17111, ThermoFisher), or with 1 nM mimic of miR-494-3p (MC12409, ThermoFisher) and 1 ng of a plasmid encoding cyclin D1 (25). Osimertinib (100 nM) or DMSO was added 24 hours post transfection. Cell viability was scored in a luminometer 72 hours post osimertinib/DMSO treatment through Cell-titer Glo cell viability assay according to manufacturer’s instruction (Promega, Cat# G7571).
Proliferation assay
Bromodeoxyuridine (BrdU) incorporation was performed according to the instructions of the commercial Fluorescein isothiocyanate (FITC) BrdU flow kit (BD Biosciences, Franklin Lakes, NJ, USA, Cat# 559619) and analyzed on a BD LSR II flow cytometer.
Apoptosis assay
Parental or osimertinib-refractory cells were seeded in triplicates, 15,000 in each replicate, in 96-well plates and treated with DMSO or with 100 nM osimertinib 24 hours post seeding. Induction of apoptosis was measured through measurement of cleaved caspase 3/7 at 24 hours post treatment, using Caspase-Glo 3/7 Assay System (Promega, Cat# G8091) according to manufacturer’s instructions.
miRNA and mRNA expression analyses
Systematic miRNA expression analysis was conducted using the nCounter Human v3 miRNA Expression Assay (NanoString Technologies, Inc., Seattle, WA, USA) covering 800 human miRNAs. Total RNA was extracted from EGFR mutant osimertinib-refractory NSCLC cell lines and EGFR mutant parental NSCLC cell lines using mirVana miRNA isolation kit (ThermoFisher, Cat# AM1560). A total of 100 ng of RNA was used as input for each sample. Hierarchical clustering and scatter plots were visualized on normalized data using nSolver analysis software. miRNA raw expression counts were normalized to housekeeping genes ACTB, B2M, GAPDH, RPL19 and RPL90. Hierarchical clustering was performed using Euclidean distance with complete linkage.
For systematic mRNA expression analysis, 100,000 parental HCC827 cells were seeded in duplicates for each experimental condition in 6-well plates and transfected, using Lipofectamine RNAiMAX (ThermoFisher), with 1 nM mimic of miR-494-3p (MC12409, ThermoFisher) or with 1 nM negative control mimic (AM17111, ThermoFisher) 24 hours post seeding. Cells were treated with 100 nM osimertinib 24 hours post transfection and harvested for total RNA isolation 48 hours post transfection. mRNA array expression was conducted using the nCounter PanCancer Pathways Panel covering 770 human cancer-associated mRNAs. RNA extraction, input and analysis were conducted as described for miRNA expression analysis.
mirVana miRNA inhibitor-library screening
miRNA inhibitor screening was conducted using a customized miRNA inhibitory library (mirVana, ThermoFisher) containing 37 experimental miRNA inhibitors as well as two control inhibitors. Parental or TKI-resistant cells were transfected with inhibitors (10 nM) in three replicates, 2,500 cells in each replicate, in 96-well plates by Lipofectamine RNAiMAX (ThermoFisher) reverse transfection. Cells were scored for viability through CellTiter-Glo cell viability assay (Promega, Cat# G7571) 72 hours post transfection. The effect on cell viability was normalized to the negative control inhibitor.
Patient sample preparation
Whole blood was drawn from 21 randomly selected NSCLC EGFR T790M-positive patients (six males, age: 72, 46, 77, 50, 63 and 56 years; 15 females, age: 56, 46, 61, 78, 74, 77, 46, 59, 83, 58, 65, 59, 73, 70 and 73 years) enrolled in the multicenter TREM-study at the Karolinska University Hospital and Oslo University Hospital, in which patients who had previously progressed on first- or second-generation EGFR TKIs were treated with osimertinib 80 mg daily (NCT02504346) (EudraCT No. 2015-000307-10) (30). Blood was collected at baseline and again at progression of disease. Plasma was separated through centrifugal isolation and aliquoted to fresh tubes and stored at −80 ℃.
Exosome RNA extraction
Exosomal RNA was isolated at the Karolinska Institutet. For each plasma/sample point, 1 mL was centrifuged at 16,000 g for 10 minutes followed by processing using the ExoRNeasy serum plasma midi kit (Qiagen, Germantown, MD, USA), as previously described (31), and the RNA was eluted in 14 µL RNase-free water.
Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) analysis
Isolated exosomal RNA was subjected to TaqMan RT-qPCR miRNA assays (ThermoFisher, Cat# 4427975) using U6 and miR-494-3p specific stem-loop primers, or to SYBR RT-qPCR using random primers (RT) and human GAPDH/cyclin D1 specific primers (qPCR) (25). Each RT reaction used 10 ng of exosomal RNA. miR-494-3p qPCR cycle threshold (Ct) values were normalized to U6 Ct values, and cyclin D1 qPCR Ct values were normalized to GAPDH Ct values, using the delta-delta-Ct method. Calculation of statistical significance was performed using a two-tailed paired t-test.
Sample definition and in-laboratory replication
All experiments, unless otherwise stated, were replicated a minimum of three times in the laboratory using biological replicates. Visualized data, unless otherwise stated, reflects a representative biological replicate. If visualized with error bars, each data point represents a minimum of three technical replicates.
Statistical analysis
Unless otherwise stated, statistical tests were performed on two-group comparisons using a two-tailed t-test, testing the area under both sides of a normal distribution. Paired t-test was performed on longitudinally sampled and matched plasma from patients, baseline versus progression of disease.
Ethical statement
The study was conducted according to the Declaration of Helsinki (as revised in 2013). The regional ethical committees, Karolinska University Hospital (registration number: 2016/944-31/1) and Oslo North Regional Ethics Board (registration number: 2015/181), approved sampling for this study. Informed consent was obtained from all subjects involved in the study.
Results
We utilized two NSCLC cell lines, NCI-H1975 and HCC827, previously generated for refractoriness to the EGFR TKI osimertinib (29). HCC827 has an exon 19 in-frame deletion in the EGFR tyrosine kinase domain (E746-A750) while NCI-H1975 harbors the double L858R/T790M mutation, rendering the latter intrinsically resistant to first-generation EGFR TKIs erlotinib and gefitinib.
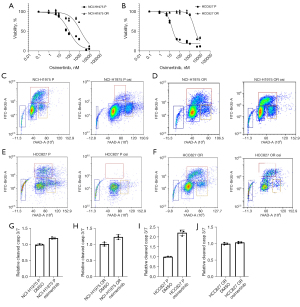
Viability of both parental lines was diminished by >10 nM TKI for 72 hours. In contrast, osimertinib-refractory lines were virtually unaffected by 10 nM TKI treatment and largely tolerated up to 1,000 nM TKI for 72 hours (Figure 1A,1B). The effect on cell viability was recapitulated when scoring parental and refractory cell lines for cell cycle progression through BrdU uptake. Parental cells entered cell cycle arrest following treatment with 100 nM TKI for 72 hours, while refractory cells continued to proliferate with minimum effects on cell cycle phase distribution (Figure 1C-1F). When analyzing parental and osimertinib-refractory cells for induction of apoptosis following treatment with osimertinib, we observed a strong induction of cleaved caspase 3/7 in parental HCC827 cells, which was completely diminished in refractory cells. NCI-H1975 cells displayed a marginal increase in early apoptosis following osimertinib treatment both in the parental and osimertinib-refractory setting (Figure 1G-1J).
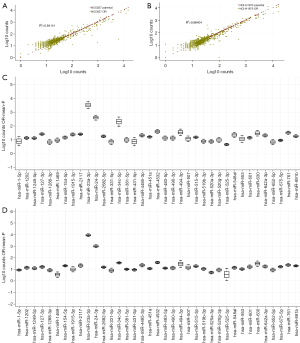
We next profiled our cell line panel for systematic miRNA expression using the hybridization platform from NanoString Technologies. No systematic alteration in expression was observed between parental and osimertinib-resistant cells (Figure 2A,2B). When investigating differential expression of individual miRNAs, refractory lines displayed blunted expression of five miRNAs, including members of the miR-141/200 family, while 37 miRNAs displayed >2-fold elevation in expression and four miRNAs displayed >3-fold elevation in expression (Figure 2C,2D, Table S1).
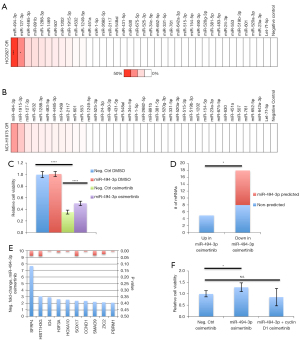
In order to investigate whether differential expression would be indicative of a functional role for specific miRNAs in conferring resistance to osimertinib, we subjected both osimertinib-refractory lines to a miRNA inhibitory library. Osimertinib-refractory cells were transiently transfected with 10 nM of 37 miRNA family inhibitors, targeting all elevated miRNAs for 72 hours. The screen revealed a substantial effect on cell viability when inhibiting miR-494-3p (Figure 3A,3B). The impact of inhibiting miR-494-3p was greater in osimertinib-resistant HCC827 cells compared to osimertinib-resistant NCI-H1975 cells, 44% reduction in cell viability and 23% reduction in cell viability, respectively, possibly reflected by the different genetic alterations leading to EGFR activation in these cell lines (Figure 3A,3B). We hypothesized that if miR-494-3p would partially confer resistance to osimertinib in osimertinib-refractory HCC827 cells, such resistance should be induced by ectopically expressing miR-494-3p in parental HCC827 cells. Ectopic expression of a miR-494-3p mimic in parental HCC827 cells alleviated the detrimental effect of osimertinib in a significant manner (Figure 3C). To obtain indications of the biological pathways affected by miR-494-3p, we extracted total RNA from osimertinib-treated parental HCC827 cells, and osimertinib-treated parental HCC827 cells ectopically expressing miR-494-3p. RNA was subjected to a pan-cancer NanoString array covering 770 cancer-associated mRNAs. Analysis revealed 18 differentially expressed transcripts (>2-fold change in expression, P<0.05) with suppressed expression in the miR-494-3p mimic + osimertinib setting compared to osimertinib alone, and five transcripts with elevated expression in the miR-494-3p mimic + osimertinib setting (Table S2). Interestingly, 10 out of 18 suppressed transcripts (SFRP4, HIST1H3G, ID4, H3F3A, HOXA10, SOX17, CCND1, SMAD9, ZIC2 and PBRM1), were found to be predicted as miR-494-3p targets using the in silico algorithm TargetScan, Release 7.2, while none of the five elevated transcripts were found to be predicted as miR-494-3p targets (Figure 3D,3E, Table S2). Kyoto encyclopedia of genes and genomes (KEGG)-term analysis resulted in several significant KEGG-pathway terms, including cell cycle and WNT signaling pathway, with cyclin D1 (CCND1) present in multiple pathways (Table S3). We have previously demonstrated cyclin D1 to be a direct functional target of miR-494-3p through genome-wide 3'UTR screens (25). Therefore, we speculated that ectopic expression of cyclin D1 might negate the impact of miR-494-3p on osimertinib in parental HCC827 cells. Ectopic expression of miR-494-3p in combination with osimertinib increased cell viability by approximately 30% compared to osimertinib alone. This effect was completely blunted when miR-494-3p was co-expressed with cyclin D1, indicative of a mechanistic relationship between miR-494-3p and cyclin D1 in modulating resistance to osimertinib (Figure 3F).
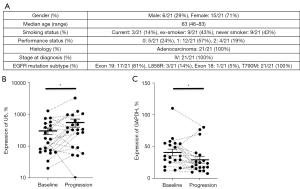
Finally, to substantiate a potential role of miR-494-3p as a marker of osimertinib-resistance, we extracted exosomal RNA from longitudinally sampled plasma at baseline and progression of disease, of 21 patients with EGFR T790M NSCLC undergoing treatment with osimertinib (Figure 4A). Extracted total RNA was subjected to stem-loop RT-qPCR miR-494-3p expression analysis and normalized to U6 snRNA. When analyzing all samples as a two-group comparison, expression of exosomal miR-494-3p was significantly elevated in progression samples compared with baseline samples. On individual sample level, plasma samples from 15 out of 21 patients displayed elevated expression of miR-494-3p at progression (Figure 4B). Moreover, we investigated the expression of cyclin D1 mRNA in the identical samples. Samples from two patients, both baseline and progression, were omitted from analysis due to failure to detect any cyclin D1 expression. In the remaining samples, and to the contrary of miR-494-3p, cyclin D1 expression was significantly reduced in progression samples compared with baseline samples. On individual sample level, plasma samples from 14 out of 19 patients displayed reduced expression of cyclin D1 at progression (Figure 4C).
Discussion
In this study we have combined miRNA profiling and inhibitory miRNA drug screening in vitro with liquid biopsy miRNA profiling in NSCLC patients to uncover miR-494-3p as a diagnostic marker as well as a potential therapeutic target in EGFR mutant NSCLC.
We found that miR-494-3p was significantly upregulated in two EGFR-mutant NSCLC cell lines refractory to the third-generation TKI osimertinib. This is one of the first reports demonstrating elevated expression of a miRNA to partially confer resistance to osimertinib. We want to emphasize that elevated expression of miR-494-3p does not solely explain the resistance mechanism to osimertinib in our cell line systems. It is possible that additional miRNAs contribute to osimertinib resistance as well. In a similar experimental setup, Li et al. (32) reported miR-184 and miR-3913-5p to be upregulated in exosomes from osimertinib-resistant cells and in plasma. miR-184 did not reach the cutoff of 2-fold elevation in expression in any of our osimertinib-resistant lines and miR-3913-5p was not included in our NanoString analysis. While these miRNAs may be important for osimertinib-resistance as well, it remains to be seen whether specific modulation of miR-184 and/or miR-3913-5p confers osimertinib resistance in vitro. Our findings may be of particular value since there are currently no obvious targeted treatment strategies for patients developing resistance to osimertinib.
Numerous reports have suggested miRNAs to play roles in human cancers, including in NSCLC (27,28,33). However, there is limited literature on miRNAs directly affecting the efficacy of EGFR TKIs in NSCLC in vitro models. The oncogenic miRNA miR-21 has been suggested to be regulated by EGFR, and to potentially substantiate the apoptotic effects of EGFR TKIs, including gefitinib in vitro (34,35). Other miRNAs reported to affect the efficacy of gefitinib, when upregulated, or downregulated, are miR-126, miR-221/222, miR-134/487b/655, and miR-19b, respectively (36-39). We did not detect any elevated expression of these miRNAs in parental versus osimertinib-refractory NSCLC cells in vitro.
In addition, the miR-30 family of miRNAs has been linked to EGFR-mutant NSCLC by several laboratories. Garofalo et al. (37) demonstrated that knockdown of EGFR in NSCLC resulted in reduced levels of miR-30b and miR-30c, and the authors proposed that these miRNAs protected NSCLC cell lines against gefitinib-induced apoptosis by targeting pro-apoptotic factors. Circulating levels of miR-30b and miR-30c were recently demonstrated to predict treatment responses to erlotinib in EGFR-mutant NSCLC patients. Low levels of miR-30b and miR-30c at start of treatment correlated with a favorable survival of EGFR-mutant NSCLC patients receiving erlotinib (40), which could potentially be linked to c-MET (37,41,42). In our systematic in vitro miRNA expression analysis, miR-30a and miR-30c were not elevated in expression in both osimertinib-refractory cell lines. It is plausible that different miRNAs play roles in the resistance to EGFR TKIs and we cannot exclude the possibility that the involvement of different miRNAs in EGFR TKI resistance may be tumor dependent, and furthermore, vary between EGFR-mutant NSCLC cell lines.
To our knowledge, there are no previous reports showing that miR-494-3p directly affects the efficacy of EGFR TKIs, and thereby functions as a modulator of EGFR TKI resistance in EGFR-driven lung cancer. However, miR-494-3p has been implicated to play roles in multiple cancer types, including lung cancer. Faversani et al. (43) reported that expression of miR-494-3p correlated with lung cancer progression and survival in both mice and in patients with KRAS mutations. Wu et al. (44) recently reported a connection between miR-494-3p and the long non-coding RNA (lncRNA) WT1-AS in NSCLC cell lines, where sponging of miR-494-3p via WT1-AS led to increased cellular apoptosis. Zhang et al. (45) reported that miR-494 modulated sensitivity to cisplatin through targeting of CASP2. Interestingly, deregulated expression of miR-494-3p was observed in cancer stem cells of chronic myeloid leukemia and correlated with resistance to the TKI imatinib (46).
Our NanoString analysis of functionally suppressed and predicted target genes following ectopic expression of miR-494-3p in combination with osimertinib resulted in significant enrichment of multiple KEGG-terms, including transforming growth factor (TGF)-beta, WNT, cell cycle and phosphatidylinositol 3-kinase (PI3K)-Akt signaling pathways. miR-494-3p has been reported to target the PI3K-Akt signaling pathway through phosphatase and tensin homolog (PTEN) in hepatocellular carcinoma, retinoblastoma and glioblastoma (47-49). Although PTEN displayed a slight downregulation following ectopic miR-494-3p expression, it did not reach our criteria for differential expression and was therefore not included in our KEGG-term analysis. However, we detected ten transcripts that in addition to being suppressed in our NanoString analysis were predicted as direct miR-494-3p targets in silico, and demonstrated that ectopic expression of cyclin D1, a previously reported target of miR-494-3p (25), negated the impact of miR-494-3p on osimertinib. Additional studies are required to understand the biological effects of cyclin D1 targeting by miR-494-3p in induced resistance to osimertinib. Since cyclin D1 expression leads to increased cellular proliferation (25), it is plausible that cyclin D1 needs to be silenced in order for cells to obtain EMT features and a higher migratory capacity, as previously reported (50).
We have uncovered miR-494-3p to partially confer resistance to osimertinib in vitro and to be significantly elevated in plasma of EGFR T790M patients progressing on osimertinib. However, the longitudinal plasma profiling was conducted on samples from a limited number of patients, which warrants validation in additional patient samples as well as in independent clinical cohorts. Nevertheless, our findings suggest that miR-494-3p may serve as a potential biomarker to monitor NSCLC disease progression on osimertinib. Moreover, given the limited therapeutic choices for patients following osimertinib resistance, it is important to further investigate the potential of targeting miR-494-3p in the in vivo setting. miRNA therapy has reached limited clinical success due to obstacles associated with delivery efficacy as well as adverse toxicities. Still, targeting miRNAs through various inhibitory approaches is advantageous due to the smaller size and rigid structure of miRNA inhibitors compared to miRNA mimic molecules (26).
In order to truly unveil the full potential of miR-494-3p in osimertinib-resistant NSCLC, it is imperative to test its therapeutic potential in in vivo models and if successful, miR-494-3p therapy may hold a future clinical promise for patients with EGFR-mutant NSCLC.
Acknowledgments
We are grateful to the staff at the KIGene core facility, Karolinska Institutet, for assisting with NanoString analysis.
Funding: This research was funded by The Swedish Cancer Society (to SE); the Sjöberg Foundation (to SE); the Swedish Research Council (to PH); the Cancer Society in Stockholm and the King Gustaf V Jubilee Fund (to SE and PH).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-21-955/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-21-955/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-21-955/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted according the Declaration of Helsinki (as revised in 2013). The regional ethical committees, Karolinska University Hospital (registration number: 2016/944-31/1) and Oslo North Regional Ethics Board (registration number: 2015/181), approved sampling for this study. Informed consent was obtained from all subjects involved in the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Dearden S, Stevens J, Wu YL, et al. Mutation incidence and coincidence in non small-cell lung cancer: meta-analyses by ethnicity and histology (mutMap). Ann Oncol 2013;24:2371-6. [Crossref] [PubMed]
- Midha A, Dearden S, McCormack R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII). Am J Cancer Res 2015;5:2892-911. [PubMed]
- Couraud S, Zalcman G, Milleron B, et al. Lung cancer in never smokers--a review. Eur J Cancer 2012;48:1299-311. [Crossref] [PubMed]
- Jänne PA, Engelman JA, Johnson BE. Epidermal growth factor receptor mutations in non-small-cell lung cancer: implications for treatment and tumor biology. J Clin Oncol 2005;23:3227-34. [Crossref] [PubMed]
- Gahr S, Stoehr R, Geissinger E, et al. EGFR mutational status in a large series of Caucasian European NSCLC patients: data from daily practice. Br J Cancer 2013;109:1821-8. [Crossref] [PubMed]
- Shi Y, Au JS, Thongprasert S, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol 2014;9:154-62. [Crossref] [PubMed]
- da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol 2011;6:49-69. [Crossref] [PubMed]
- Morgillo F, Della Corte CM, Fasano M, et al. Mechanisms of resistance to EGFR-targeted drugs: lung cancer. ESMO Open 2016;1:e000060. [Crossref] [PubMed]
- Coudert B, Ciuleanu T, Park K, et al. Survival benefit with erlotinib maintenance therapy in patients with advanced non-small-cell lung cancer (NSCLC) according to response to first-line chemotherapy. Ann Oncol 2012;23:388-94. [Crossref] [PubMed]
- Chang A, Parikh P, Thongprasert S, et al. Gefitinib (IRESSA) in patients of Asian origin with refractory advanced non-small cell lung cancer: subset analysis from the ISEL study. J Thorac Oncol 2006;1:847-55. [Crossref] [PubMed]
- Yang JC, Wu YL, Schuler M, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol 2015;16:141-51. [Crossref] [PubMed]
- Ramalingam SS, Jänne PA, Mok T, et al. Dacomitinib versus erlotinib in patients with advanced-stage, previously treated non-small-cell lung cancer (ARCHER 1009): a randomised, double-blind, phase 3 trial. Lancet Oncol 2014;15:1369-78. [Crossref] [PubMed]
- Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2005;2:e73. [Crossref] [PubMed]
- Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005;352:786-92. [Crossref] [PubMed]
- Suda K, Onozato R, Yatabe Y, et al. EGFR T790M mutation: a double role in lung cancer cell survival? J Thorac Oncol 2009;4:1-4. [Crossref] [PubMed]
- Bell DW, Gore I, Okimoto RA, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet 2005;37:1315-6. [Crossref] [PubMed]
- Chen HJ, Mok TS, Chen ZH, et al. Clinicopathologic and molecular features of epidermal growth factor receptor T790M mutation and c-MET amplification in tyrosine kinase inhibitor-resistant Chinese non-small cell lung cancer. Pathol Oncol Res 2009;15:651-8. [Crossref] [PubMed]
- Schuler M, Yang JC, Park K, et al. Afatinib beyond progression in patients with non-small-cell lung cancer following chemotherapy, erlotinib/gefitinib and afatinib: phase III randomized LUX-Lung 5 trial. Ann Oncol 2016;27:417-23. [Crossref] [PubMed]
- Lamb YN, Scott LJ. Osimertinib: A Review in T790M-Positive Advanced Non-Small Cell Lung Cancer. Target Oncol 2017;12:555-62. [Crossref] [PubMed]
- Ramalingam SS, Vansteenkiste J, Planchard D, et al. Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N Engl J Med 2020;382:41-50. [Crossref] [PubMed]
- Jänne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015;372:1689-99. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-33. [Crossref] [PubMed]
- Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature 2005;435:834-8. [Crossref] [PubMed]
- Hydbring P, Wang Y, Bogorad RL, et al. Identification of cell cycle-targeting microRNAs through genome-wide screens. Cell Cycle 2017;16:2241-8. [Crossref] [PubMed]
- Hydbring P, Wang Y, Fassl A, et al. Cell-Cycle-Targeting MicroRNAs as Therapeutic Tools against Refractory Cancers. Cancer Cell 2017;31:576-90.e8. [Crossref] [PubMed]
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16:203-22. [Crossref] [PubMed]
- Hydbring P, Badalian-Very G. Clinical applications of microRNAs. F1000Res 2013;2:136. [Crossref] [PubMed]
- Lai Y, Kacal M, Kanony M, et al. miR-100-5p confers resistance to ALK tyrosine kinase inhibitors Crizotinib and Lorlatinib in EML4-ALK positive NSCLC. Biochem Biophys Res Commun 2019;511:260-5. [Crossref] [PubMed]
- McGowan M, Kleinberg L, Halvorsen AR, et al. NSCLC depend upon YAP expression and nuclear localization after acquiring resistance to EGFR inhibitors. Genes Cancer 2017;8:497-504. [Crossref] [PubMed]
- Eide IJZ, Helland Å, Ekman S, et al. Osimertinib in T790M-positive and -negative patients with EGFR-mutated advanced non-small cell lung cancer (the TREM-study). Lung Cancer 2020;143:27-35. [Crossref] [PubMed]
- Enderle D, Spiel A, Coticchia CM, et al. Characterization of RNA from Exosomes and Other Extracellular Vesicles Isolated by a Novel Spin Column-Based Method. PLoS One 2015;10:e0136133. [Crossref] [PubMed]
- Li X, Chen C, Wang Z, et al. Elevated exosome-derived miRNAs predict osimertinib resistance in non-small cell lung cancer. Cancer Cell Int 2021;21:428. [Crossref] [PubMed]
- Langsch S, Baumgartner U, Haemmig S, et al. miR-29b Mediates NF-κB Signaling in KRAS-Induced Non-Small Cell Lung Cancers. Cancer Res 2016;76:4160-9. Erratum in: Cancer Res 2016;76:6436. [Crossref] [PubMed]
- Seike M, Goto A, Okano T, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci U S A 2009;106:12085-90. [Crossref] [PubMed]
- Li B, Ren S, Li X, et al. MiR-21 overexpression is associated with acquired resistance of EGFR-TKI in non-small cell lung cancer. Lung Cancer 2014;83:146-53. [Crossref] [PubMed]
- Zhong M, Ma X, Sun C, et al. MicroRNAs reduce tumor growth and contribute to enhance cytotoxicity induced by gefitinib in non-small cell lung cancer. Chem Biol Interact 2010;184:431-8. [Crossref] [PubMed]
- Garofalo M, Romano G, Di Leva G, et al. EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat Med 2011;18:74-82. [Crossref] [PubMed]
- Kitamura K, Seike M, Okano T, et al. MiR-134/487b/655 cluster regulates TGF-β-induced epithelial-mesenchymal transition and drug resistance to gefitinib by targeting MAGI2 in lung adenocarcinoma cells. Mol Cancer Ther 2014;13:444-53. [Crossref] [PubMed]
- Baumgartner U, Berger F, Hashemi Gheinani A, et al. miR-19b enhances proliferation and apoptosis resistance via the EGFR signaling pathway by targeting PP2A and BIM in non-small cell lung cancer. Mol Cancer 2018;17:44. [Crossref] [PubMed]
- Hojbjerg JA, Ebert EBF, Clement MS, et al. Circulating miR-30b and miR-30c predict erlotinib response in EGFR-mutated non-small cell lung cancer patients. Lung Cancer 2019;135:92-6. [Crossref] [PubMed]
- Cappuzzo F, Jänne PA, Skokan M, et al. MET increased gene copy number and primary resistance to gefitinib therapy in non-small-cell lung cancer patients. Ann Oncol 2009;20:298-304. [Crossref] [PubMed]
- Leonetti A, Sharma S, Minari R, et al. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer 2019;121:725-37. [Crossref] [PubMed]
- Faversani A, Amatori S, Augello C, et al. miR-494-3p is a novel tumor driver of lung carcinogenesis. Oncotarget 2017;8:7231-47. [Crossref] [PubMed]
- Wu C, Yang J, Li R, et al. LncRNA WT1-AS/miR-494-3p Regulates Cell Proliferation, Apoptosis, Migration and Invasion via PTEN/PI3K/AKT Signaling Pathway in Non-Small Cell Lung Cancer. Onco Targets Ther 2021;14:891-904. [Crossref] [PubMed]
- Zhang Q, Li Y, Zhao M, et al. MiR-494 acts as a tumor promoter by targeting CASP2 in non-small cell lung cancer. Sci Rep 2019;9:3008. [Crossref] [PubMed]
- Salati S, Salvestrini V, Carretta C, et al. Deregulated expression of miR-29a-3p, miR-494-3p and miR-660-5p affects sensitivity to tyrosine kinase inhibitors in CML leukemic stem cells. Oncotarget 2017;8:49451-69. [Crossref] [PubMed]
- Lin H, Huang ZP, Liu J, et al. MiR-494-3p promotes PI3K/AKT pathway hyperactivation and human hepatocellular carcinoma progression by targeting PTEN. Sci Rep 2018;8:10461. [Crossref] [PubMed]
- Li XT, Wang HZ, Wu ZW, et al. miR-494-3p Regulates Cellular Proliferation, Invasion, Migration, and Apoptosis by PTEN/AKT Signaling in Human Glioblastoma Cells. Cell Mol Neurobiol 2015;35:679-87. [Crossref] [PubMed]
- Xu F, Liu G, Wang L, et al. miR-494 promotes progression of retinoblastoma via PTEN through PI3K/AKT signaling pathway. Oncol Lett 2020;20:1952-60. [Crossref] [PubMed]
- Tobin NP, Sims AH, Lundgren KL, et al. Cyclin D1, Id1 and EMT in breast cancer. BMC Cancer 2011;11:417. [Crossref] [PubMed]


