
Tumor volume as a predictor of cell free DNA mutation detection in advanced non-small cell lung cancer
Introduction
Lung cancer remains the most common cause of cancer-associated death in the United States (1). There is substantial unmet need for biomarkers to guide therapeutic interventions in lung cancer (2). Cell free DNA (cfDNA) in blood plasma is a biomarker that allows evaluation of genetic alterations in cancer, and its utility as a tool to direct therapeutic interventions in lung cancer is increasing (3). Most cfDNA in the blood stream arises from hematopoietic cell lineages (4). Circulating tumor DNA (ctDNA) is the subset of cfDNA that is derived from tumor tissue (5). Shedding of ctDNA is thought to be from apoptosis, necrosis, or active secretion of cancer cells (6-8). Analysis of plasma cfDNA using next generation sequencing (NGS) is an effective, non-invasive method of detecting actionable genetic mutations in lung cancer patients, and has high concordance to mutations detected by tissue analysis along with a faster test turn-around time (3).
Factors that determine ctDNA shedding dynamics remain an active area of research. Tumor burden, assessed in a variety of ways, has frequently been found to be associated with ctDNA. Across a broad range of primary cancer sites, tumor stage has been shown to be associated with detection of cfDNA mutations (9-11). Total tumor volume has been shown to be associated with amount of overall cfDNA (i.e., tumor and non-tumor derived) in patients with lung cancer (12) and melanoma (13) as well as with amount of ctDNA in lung cancer (14) and melanoma (15). However, tumor volume assessment has often relied upon surrogates such as calculated volume based on measurement of cross-sectional dimensions (13-16), which may provide an incomplete picture of total tumor volume.
Other possible contributors to ctDNA shedding dynamics include the tissue in which a tumor or metastasis is located, tumor metabolism, and tumor histology. Data on early cancer detection has found differential rates of cancer detection based on primary tumor site, with lower detection rates in sites such as the central nervous system (CNS) and higher detection rates in sites such as the liver (10,11), and there may be tissue-specific factors that contribute to ctDNA shedding (17). Evaluation of patients with metastatic CNS disease have also shown lower levels of plasma ctDNA in those populations (18,19). Conversely, higher levels of ctDNA have been found in patients with metastases to bone (18,20) and viscera (18). Tumor metabolic activity has also been shown to correlate with cfDNA (20) and ctDNA (16,21) levels. Tumor histology has been found to be associated with ctDNA detection in early stage NSCLC with squamous histology associated with higher rates of ctDNA detection (22).
Furthermore, the presence of baseline detectable ctDNA is associated with increased risk of recurrence and decreased overall survival (OS) across a variety of primary tumor sites, including lung (21,23), colorectal (24), breast (25), and melanoma (15), and baseline level of Epstein-Barr viral DNA is associated with worse OS in nasopharynx cancer (26,27). However, the interaction between ctDNA detection and tumor volume in association with survival has not been well explored.
We performed a detailed quantitative analysis of tumor volume in patients with non-small cell lung cancer (NSCLC) treated at our institution who underwent plasma cfDNA NGS testing to evaluate the association of tumor volume with cfDNA mutation detection and OS. We present the following article in accordance with the REMARK reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-164/rc).
Methods
Patient population
Patients with advanced NSCLCs were prospectively enrolled on an institutional review board-approved plasma NGS genotyping protocol, as previously described (3). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Institutional Review Board of Memorial Sloan Kettering Cancer Center (Protocol 12-245) and informed consent was taken from all individual participants. Enrollment was continuous, not blinded, and not randomized. All patients had radiographic evidence of disease, were age 18 or older, and signed written informed consent. Patient sex of male and female was recorded and balanced research subject participation was encouraged. There were 459 patients enrolled from 11/2016 to 8/2018. Patients in the present analysis were required to have stage III or IV NSCLC and be treatment naïve at the time of plasma cfDNA lab draw since treatment would be expected to bias mutation detection. Any patients who lacked adequate baseline imaging for volumetric analysis within 60 days of cfDNA draw were excluded. There were 110 patients who met inclusion criteria. Treatment was physician’s choice and not randomized.
Tumor volumetric analysis
All patients underwent PET/CT and MRI brain imaging at diagnosis, and these images were used for volumetric analysis. Imaging was transferred from PACS into a radiation oncology contouring software package for volume delineation (MIM, Beachwood, OH, USA). Volume of tumor was separately delineated by a blinded reviewer for the following anatomical locations: lung, pleura, lymph nodes, brain, bone, liver, adrenal, other viscera, soft tissue, and other. Volume delineation was performed manually based on careful visual inspection of both PET and CT images, as well as brain MRI when appropriate, and correlation with clinical radiology reports. Select cases were audited by a second observer for consistency. For analysis, total tumor volume was the sum of tumor volume from all anatomical locations, intrathoracic tumor volume was the sum of tumor volume from lung, pleura, and lymph nodes, and visceral tumor volume was the sum of tumor volume from liver, adrenal, and other viscera. Tumor volume in the brain was delineated based on T1 post-contrast imaging. Tumor volume in the body was delineated using a combination of attenuation corrected PET and associated diagnostic CT images.
Plasma NGS genotyping
Plasma NGS genotyping was performed as previously described (3). Briefly, peripheral venous blood was collected using standard phlebotomy techniques into two Streck tubes (Omaha, NE; 10 mL each). Streck tubes were shipped at room temperature using overnight express mail to Resolution Bioscience (ResBio, Redmond, WA). DNA was extracted in a Clinical Laboratory Improvement Amendments certified laboratory. Targeted NGS was performed using a validated, bias-corrected, hybrid-capture assay of 21-genes (ResBio ctDx-Lung) with median unique reads of 3,000× and sensitive detection at variant allele frequency (VAF) above 0.1% (28). Genes included in the panel are: AKT1, ALK, BRAF, EGFR, FGFR1, FGFR2, FGFR3, HER2 (ERBB2), JAK2, KRAS, NRAS, NTRK1, MAP2K1, MET, MYC, CD274, PIK3CA, RET, RICTOR, ROS1, TP53. The study investigators were provided with detailed reports of observed point mutations, insertions, deletions, gene fusions, and copy number alterations in a secure, online repository.
Tissue genotyping
All patients underwent NGS of tissue biopsies, which was performed using a hybridization capture assay, Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT), as previously described (3,29). Mutations were reviewed across plasma and tissue NGS results to determine concordance. Concordance was defined as patients where at least one identical genomic alteration was found in both tissue and plasma.
Outcome measures and variables
The primary endpoint was presence of a detectable mutation on the plasma NGS assay, assessed as a binary outcome. Secondary endpoints included OS and VAF. In cases where more than one mutation was detected, the largest VAF was used, and no VAF threshold was used to filter results.
Variables used in the models of cfDNA mutation detection and VAF included total tumor volume (binary and continuous), lung tumor volume, intrathoracic tumor volume, bone tumor volume, viscera tumor volume, brain tumor volume, volume per tumor, presence of tumor in bone, presence of tumor in viscera, and presence of tumor in brain. Variables used in the model of OS included presence of cfDNA mutation detection, total tumor volume (binary and continuous), lung tumor volume, intrathoracic tumor volume, bone tumor volume, viscera tumor volume, brain tumor volume, volume per tumor, presence of tumor in bone, presence of tumor in viscera, and presence of tumor in brain.
Statistical analysis
Continuous variables were described using median and ranges, whereas categorical variables were summarized using frequency and percentages. The maxstat package was used to determine an optimal cut-point for total tumor volume, and demographic and clinical variables were assessed for differences based on this cut-point using Wilcoxon rank sum, Chi-squared, and Fisher’s exact tests. Univariate logistic regression was utilized to find variables associated with the presence or absence of cfDNA mutation, and then a multivariable model was built using variables that showed significance univariately. OS was defined as time from cfDNA plasma draw until death or last follow-up and was assessed using Kaplan-Meier methodology. Univariate Cox proportional hazard models were built to find factors associated with OS, and significant variables were included in a multivariable OS model. Univariate linear regression was used to evaluate association between variables of interest and VAF. Plasma mutation genes are reported descriptively both for the overall cohort, and by above/below the cut-point threshold. All analyses were done in R v4.0.5.
Results
Patient characteristics, mutations, and imaging
A total of 110 patients were included in the present analysis. Most patients were female (62/110, 56%), White (83/110, 78%), former smokers (67/110, 61%), and had adenocarcinoma histology (93/110, 85%) (Table 1). There were 104 patients with metastatic disease (95%). There were significantly more patients with visceral metastases in the group with a total tumor volume greater than 65 mL (26/58, 45% vs. 10/52, 19%; P=0.004), and there was a trend toward more patients with bone metastases in the group with a total tumor volume greater than 65 mL (32/58, 55% vs. 20/52, 38%; P=0.080). There were 12 patients with metastases in both bone and liver. There were numerically more patients with “Other” histology in the group with total tumor volume greater than 65 mL, but this did not reach statistical significance (8/58, 14% vs. 2/52, 4%; P=0.10). Median follow up among alive patients was 38 months.
Table 1
| Characteristic | Overall (N=110) | TTV ≤65 mL (N=52) | TTV >65 mL (N=58) | P value* |
|---|---|---|---|---|
| Age (years) | 68 [28, 91] | 67 [28, 91] | 69 [35, 89] | 0.31 |
| Sex | 0.61 | |||
| Female | 62 (56%) | 28 (54%) | 34 (59%) | |
| Male | 48 (44%) | 24 (46%) | 24 (41%) | |
| Race | 0.010 | |||
| White | 83 (78%) | 38 (76%) | 45 (79%) | |
| Asian | 17 (16%) | 12 (24%) | 5 (9%) | |
| Black | 5 (5%) | 0 (0%) | 5 (9%) | |
| Other | 2 (2%) | 0 (0%) | 2 (4%) | |
| Smoking status | 0.37 | |||
| Former smoker | 67 (61%) | 29 (56%) | 38 (66%) | |
| Current smoker | 1 (1%) | 1 (2%) | 0 (0%) | |
| Never smoker | 42 (38%) | 22 (42%) | 20 (34%) | |
| Pack years | 26 [1, 82] | 22 [1, 82] | 30 [1, 80] | 0.12 |
| Unknown | 44 | 23 | 21 | |
| Histology | 0.20 | |||
| Adenocarcinoma | 93 (85%) | 47 (90%) | 46 (79%) | |
| Squamous cell carcinoma | 7 (6%) | 3 (6%) | 4 (7%) | |
| Other | 10 (9%) | 2 (4%) | 8 (14%) | |
| Histology collapsed | 0.10 | |||
| Adeno/SCC | 100 (91%) | 50 (96%) | 50 (86%) | |
| Other | 10 (9%) | 2 (4%) | 8 (14%) | |
| Bone tumor present | 0.080 | |||
| No | 58 (53%) | 32 (62%) | 26 (45%) | |
| Yes | 52 (47%) | 20 (38%) | 32 (55%) | |
| Viscera tumor present | 0.004 | |||
| No | 74 (67%) | 42 (81%) | 32 (55%) | |
| Yes | 36 (33%) | 10 (19%) | 26 (45%) | |
| Liver tumor present | 0.28 | |||
| No | 93 (85%) | 46 (88%) | 47 (81%) | |
| Yes | 17 (15%) | 6 (12%) | 11 (19%) | |
| Brain tumor present | 0.87 | |||
| No | 77 (70%) | 36 (69%) | 41 (71%) | |
| Yes | 33 (30%) | 16 (31%) | 17 (29%) | |
| KRAS mutation in plasma | 0.18 | |||
| No | 57 (73%) | 23 (82%) | 34 (68%) | |
| Yes | 21 (27%) | 5 (18%) | 16 (32%) | |
| EGFR mutation in plasma | 0.65 | |||
| No | 59 (76%) | 22 (79%) | 37 (74%) | |
| Yes | 19 (24%) | 6 (21%) | 13 (26%) | |
| ALK mutation in plasma | 0.48 | |||
| No | 67 (86%) | 23 (82%) | 44 (88%) | |
| Yes | 11 (14%) | 5 (18%) | 6 (12%) | |
| TP53 mutation in plasma | 0.97 | |||
| No | 67 (86%) | 24 (86%) | 43 (86%) | |
| Yes | 11 (14%) | 4 (14%) | 7 (14%) | |
| Other plasma mutation genes | ||||
| BRAF | 2 (3%) | 1 (4%) | 1 (2%) | |
| ERBB2 | 1 (1%) | 0 (0%) | 1 (2%) | |
| FGFR1 | 1 (1%) | 1 (4%) | 0 (0%) | |
| MET | 8 (10%) | 5 (18%) | 3 (6%) | |
| NRAS | 1 (1%) | 1 (4%) | 0 (0%) | |
| RET | 2 (3%) | 0 (0%) | 2 (4%) | |
| ROS1 | 1 (1%) | 0 (0%) | 1 (2%) | |
| None detected | 32 (29%) | 24 (46%) | 8 (14%) |
The data are shown as median [range] or n (%); *, Wilcoxon rank sum test; Pearson’s Chi-squared test; Fisher’s exact test. TTV, total tumor volume; Adeno, adenocarcinoma; SCC, squamous cell carcinoma.
There were 78 (71%) patients with a mutation detected on plasma NGS. Most patients had 1 mutation detected (n=40), while there were 26, 9, and 3 patients with 2, 3, and 4+ mutations detected, respectively. The genes with the highest VAF mutation for each patient with a mutation detected are listed in Table 1 and depicted in Figure S1. The most commonly mutated genes were KRAS (21/78, 27%), EGFR (19/78, 24%), ALK (11/78, 14%), and TP53 (11/78, 14%). The most common specific mutations were EGFR L858R (n=9) and KRAS G12C (n=7).
All 110 patients had corresponding tissue NGS results, 89 patients had a mutation detected on tissue NGS, and 52.7% (58/110; 95% CI: 43.0–62.2%) had concordant results between tissue and plasma. Among patients who tested positive on plasma NGS, 74.4% (58/78; 95% CI: 63.0–83.3%) were concordant on tissue NGS. Among patients who tested positive on tissue NGS, 65.2% (58/89; 95% CI: 54.3–74.8%) were concordant on plasma NGS.
The median total tumor volume was 66 mL (range, 2–1,383 mL) (Table 2). The most common extrathoracic metastatic site was bone (52/110, 47%), and this was also the extrathoracic site with greatest median tumor volume (13 mL, range, 0.3–278 mL). The median time difference between plasma draw and imaging was 13 days (IQR, 5–27 days). Detailed, per-patient volumetric data and mutation detection data is provided in Table S1.
Table 2
| Characteristic | N | Median [range] (mL) |
|---|---|---|
| Total tumor volume | 110 | 66 [2–1,383] |
| Lung tumor volume | 108 | 21 [0.4–347] |
| Pleura tumor volume | 26 | 4 [0.4–240] |
| Lymph node tumor volume | 93 | 10 [0.2–651] |
| Intrathoracic tumor volume (lung, pleura and lymph node) | 110 | 40 [2–994] |
| Liver tumor volume | 17 | 6 [0.3–307] |
| Adrenal tumor volume | 19 | 2.2 [0.2–34.3] |
| Other viscera tumor volume | 6 | 25 [0.1–84] |
| All viscera tumor volume (liver, adrenal and other viscera) | 36 | 5 [0.1–309] |
| Bone tumor volume | 52 | 13 [0.3–278] |
| Brain tumor volume | 33 | 2.0 [0.1–38.5] |
| Soft tissue tumor volume | 15 | 5 [0.4–376] |
| Other tumor volume | 4 | 7.4 [0.2–12.3] |
Tumor volume and predictors of cfDNA mutation detection
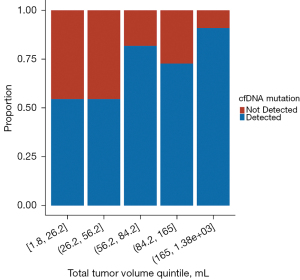
Median total tumor volume for patients with a cfDNA mutation detected was 76 mL (range, 5–1,383 mL) compared to 45 mL (range, 2–460 mL) for patients with no mutation detected (P=0.002; Wilcoxon rank sum). A segmented bar chart was generated to evaluate the relationship between total tumor volume and proportion of patients with cfDNA mutation detected (Figure 1), which depicts a step function increase in proportion of patients with a detection event at higher total tumor volumes. Cut-point analysis was performed and identified the optimal total tumor volume threshold to be associated with cfDNA mutation detection, which was 64.7 mL; above this value the probability of a mutation detection event was significantly higher (P=0.006).
Univariate analysis was performed to evaluate factors associated with presence or absence of cfDNA mutation detection (Table 3). Variables found to have a significant association with cfDNA mutation detection included total tumor volume (continuous variable; OR: 1.07, 95% CI: 1.01–1.14, P=0.028), total tumor volume (binary variable using 65 mL threshold; OR: 5.36, 95% CI: 2.20–14.2, P<0.001), presence of bone metastases (OR: 3.14, 95% CI: 1.32–7.97, P=0.012), presence of visceral metastases (OR: 3.56, 95% CI: 1.32–11.4, P=0.018) and being a never smoker (OR: 0.42, 95% CI: 0.18–0.96, P=0.041). Of note, no subsite tumor volume was significantly associated with cfDNA mutation detection.
Table 3
| Characteristic | N | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P value | OR | 95% CI | P value | |||
| Age | 110 | 1.00 | 0.97, 1.03 | 0.98 | ||||
| Sex | ||||||||
| Female | 62 | – | – | |||||
| Male | 48 | 0.70 | 0.30, 1.59 | 0.39 | ||||
| Smoking status | ||||||||
| Current/former smoker | 68 | – | – | – | – | |||
| Never smoker | 42 | 0.42 | 0.18, 0.96 | 0.041 | 0.40 | 0.15, 1.02 | 0.057 | |
| Histology | ||||||||
| Adenocarcinoma | 93 | – | – | |||||
| Other | 10 | 1.64 | 0.38, 11.3 | 0.55 | ||||
| Squamous cell carcinoma | 7 | 0.55 | 0.11, 2.92 | 0.45 | ||||
| Total tumor volume (binary) | ||||||||
| ≤65 mL | 52 | – | – | – | – | |||
| >65 mL | 58 | 5.36 | 2.20, 14.2 | <0.001 | 4.30 | 1.70, 11.8 | 0.003 | |
| Total tumor volume (continuous) | 110 | 1.07 | 1.01, 1.14 | 0.028 | ||||
| Lung tumor volume | 108 | 1.06 | 0.99, 1.17 | 0.13 | ||||
| Intrathoracic tumor volume | 110 | 1.04 | 1.00, 1.11 | 0.14 | ||||
| Bone tumor volume | 52 | 1.10 | 0.94, 1.45 | 0.37 | ||||
| Viscera tumor volume | 36 | 1.00 | 0.87, 1.30 | 0.99 | ||||
| Liver tumor volume | 17 | 1.20 | 0.93, 3.29 | 0.48 | ||||
| Brain tumor volume | 33 | 0.78 | 0.31, 2.17 | 0.58 | ||||
| Volume per tumor | 110 | 0.96 | 0.82, 1.11 | 0.50 | ||||
| Bone tumor present | ||||||||
| No | 58 | – | – | – | – | |||
| Yes | 52 | 3.14 | 1.32, 7.97 | 0.012 | 2.68 | 1.06, 7.23 | 0.042 | |
| Viscera tumor present | ||||||||
| No | 74 | – | – | – | – | |||
| Yes | 36 | 3.56 | 1.32, 11.4 | 0.018 | 2.47 | 0.83, 8.42 | 0.12 | |
| Liver tumor present | ||||||||
| No | 93 | – | – | |||||
| Yes | 17 | 1.40 | 0.45, 5.31 | 0.58 | ||||
| Brain tumor present | ||||||||
| No | 77 | – | – | |||||
| Yes | 33 | 1.42 | 0.57, 3.76 | 0.46 | ||||
cfDNA, cell free DNA; OR, odds ratio; CI, confidence interval.
Total tumor volume as a binary variable was carried forward into multivariate analysis along with presence of bone metastases, presence of visceral metastases, and smoking status (Table 3). Total tumor volume remained significant as a predictor of detection of cfDNA mutation (OR: 4.30, 95% CI: 1.70–11.8, P=0.003), as did presence of bone metastases (OR: 2.68, 95% CI: 1.06–7.23, P=0.042). Never smoker status trended toward significance (OR: 0.40, 95% CI: 0.15–1.02, P=0.057).
Additional analysis was performed among patients with a mutation detection event to assess for any differences in baseline patient and tumor characteristics between patients with total tumor volume greater than versus less than or equal to the cut point of 65 mL. The number of patients with total tumor volume of 65 mL or less versus greater than 65 mL were 28 and 50, respectively. Variables assessed included age, sex, race, smoking status, smoking pack years, tumor histology, and presence or absence of tumor in tissue sites of bone, brain, or viscera. There were significantly fewer patients with total tumor volume of 65 mL or less who had a visceral tumor (21%, 6/28) compared with total tumor volume greater than 65 mL (50%, 25/50; P=0.013). No other differences were found.
Predictors of OS
Univariate analysis of OS found factors having a significant association with OS included age (HR: 1.03, 95% CI: 1.01–1.05, P=0.005), total tumor volume (continuous variable; HR: 1.01, 95% CI: 1.00–1.02, P=0.016), total tumor volume (binary variable using 65 mL threshold; HR: 2.10, 95% CI: 1.29–3.42, P=0.003), volume per tumor (HR: 1.13, 95% CI: 1.03–1.24, P=0.011), detection of cfDNA mutation (HR: 2.62, 95% CI: 1.43–4.79, P=0.002), presence of bone metastases (HR: 1.67, 95% CI: 1.04–2.68, P=0.032), presence of visceral metastases (HR: 1.84, 95% CI: 1.14–2.97, P=0.013), and presence of liver metastases (HR: 2.08, 95% CI: 1.17–3.69, P=0.012) (Table 4).
Table 4
| Characteristic | N | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |||
| Age | 110 | 1.03 | 1.01, 1.05 | 0.005 | 1.04 | 1.01, 1.06 | 0.002 | |
| Sex | ||||||||
| Female | 62 | – | – | |||||
| Male | 48 | 1.02 | 0.63, 1.64 | 0.95 | ||||
| Smoking status | ||||||||
| Current/former smoker | 68 | – | – | |||||
| Never smoker | 42 | 0.75 | 0.46, 1.23 | 0.26 | ||||
| Histology | ||||||||
| Adenocarcinoma | 93 | – | – | |||||
| Other | 10 | 1.06 | 0.46, 2.47 | 0.89 | ||||
| Squamous cell carcinoma | 7 | 1.46 | 0.63, 3.39 | 0.38 | ||||
| Presence of cfDNA mutation detection | 110 | 2.62 | 1.43, 4.79 | 0.002 | 2.11 | 1.10, 4.03 | 0.024 | |
| Total tumor volume (binary) | ||||||||
| ≤65 mL | 52 | – | – | – | – | |||
| >65 mL | 58 | 2.10 | 1.29, 3.42 | 0.003 | 1.60 | 0.94, 2.71 | 0.084 | |
| Total tumor volume (continuous) | 110 | 1.01 | 1.00, 1.02 | 0.016 | ||||
| Lung tumor volume | 108 | 1.01 | 0.98, 1.04 | 0.42 | ||||
| Intrathoracic tumor volume | 110 | 1.01 | 1.00, 1.03 | 0.13 | ||||
| Bone tumor volume | 52 | 1.03 | 0.98, 1.09 | 0.21 | ||||
| Viscera tumor volume | 36 | 1.02 | 0.96, 1.08 | 0.48 | ||||
| Liver tumor volume | 17 | 1.02 | 0.95, 1.08 | 0.63 | ||||
| Brain tumor volume | 33 | 1.00 | 0.60, 1.66 | 0.99 | ||||
| Volume per tumor | 110 | 1.13 | 1.03, 1.24 | 0.011 | ||||
| Bone tumor present | – | – | ||||||
| No | 58 | – | – | 1.66 | 1.01, 2.73 | 0.047 | ||
| Yes | 52 | 1.67 | 1.04, 2.68 | 0.032 | ||||
| Viscera tumor present | ||||||||
| No | 74 | – | – | – | – | |||
| Yes | 36 | 1.84 | 1.14, 2.97 | 0.013 | 0.90 | 0.47, 1.73 | 0.75 | |
| Liver tumor present | ||||||||
| No | 93 | – | – | – | – | |||
| Yes | 17 | 2.08 | 1.17, 3.69 | 0.012 | 2.12 | 0.99, 4.53 | 0.053 | |
| Brain tumor present | ||||||||
| No | 77 | – | – | |||||
| Yes | 33 | 0.60 | 0.35, 1.03 | 0.065 | ||||
OR, odds ratio; CI, confidence interval; cfDNA, cell free DNA.
Factors included in multivariate analysis were age, total tumor volume (binary), detection of cfDNA mutation, presence of bone metastases, presence of visceral metastases, and presence of liver metastases. Factors that remained significant as predictors of OS included age (HR: 1.04, 95% CI: 1.01–1.06, P=0.002), detection of cfDNA mutation (HR: 2.11, 95% CI: 1.10–4.03, P=0.024), and presence of bone metastases (HR: 1.66, 95% CI: 1.01–2.73, P=0.047) (Table 4). Total tumor volume (HR: 1.60, 95% CI: 0.94–2.71, P=0.084) and presence of liver metastases (HR: 2.12, 95% CI: 0.99–4.53, P=0.053) trended toward significance. Kaplan-Meier curves were generated illustrating OS according to the three variables that remained significant on multivariate analysis (Figure 2A-2C).
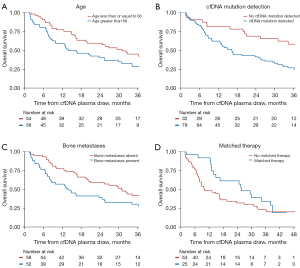
We additionally analyzed survival among patients with a mutation present according to whether the mutation was matched to a therapeutic target or not. Patients with a matched therapy had a median OS of 26 months, compared with 10 months for patients with a mutation that was not matched to a therapy, although this difference was not significant (Figure 2D; HR 0.62; 95% CI: 0.35–1.10, P=0.10).
Predictors of VAF
Additional analysis was performed to assess variables that may be associated with VAF. Univariate analysis including the same factors used in the primary endpoint analysis was performed. No variables were found to be significantly associated with VAF (data not shown).
Discussion
To our knowledge, this is the first clinical study to perform detailed tumor volume delineation and correlate it with plasma cfDNA NGS findings in patients with advanced NSCLC. We identified a total tumor volume threshold above which the probability of detecting a mutation in plasma cfDNA was much more likely. We furthermore showed that total tumor volume above the threshold of 65 mL was significantly associated with OS on univariate analysis and trended toward significance on multivariate analysis.
In this study, we were primarily interested in exploring factors that impact shedding of ctDNA. In particular, we investigated two separate but related lines of inquiry. The first is whether there is an association between ctDNA shedding and the presence of cancer metastases in certain organ systems. On univariate analysis, we found an association between metastases in bone and viscera and mutation detection, which is consistent with prior reports in patients with melanoma (18) and lung cancer (20). On multivariate analysis, presence of bone metastasis remained significant as a predictor of mutation detection (P=0.042), which may indicate that metastases in bone have a greater propensity to shed ctDNA than other tissue sites.
Our second line of inquiry is on the nature of the relationship between ctDNA shedding dynamics and volume of tumor, either total tumor volume or volume in a particular organ system or subsite. As expected, we found that patients with a mutation detected had larger total tumor volumes. Total tumor volume as a continuous variable was associated with mutation detection, but we did not find an association between mutation detection and any tumor volume subsites. In particular, while presence of bone metastasis was associated with mutation detection, volume of bone metastasis was not, even though there was a broad range of bone metastasis volumes. This may indicate that metastases to bone are generally more likely to shed ctDNA but that the threshold for bone metastasis volume to produce this effect is small. To further explore total tumor volume, we generated a bar chart to evaluate its relationship to mutation detection. The resulting Figure 1 demonstrated a step increase in proportion of patients with a mutation detection event above a certain threshold volume. The bar chart provided strong rationale for subsequent analysis showing that the optimal threshold cut-point for increased likelihood of mutation detection was 65 mL.
When analyzing the relationship between total tumor volume as a binary variable with the identified 65 mL volume threshold, we found a much higher odds ratio for mutation detection compared with analysis of total tumor volume as a continuous variable (OR 5.36 vs. 1.07). In combination with the bar chart findings, this led us to proceed with the binary representation of total tumor volume in the multivariate analysis, which showed that total tumor volume greater than 65 mL was the most significant factor, and had the greatest effect size magnitude, in predicting likelihood of mutation detection. Based on these results, it may be the case that shedding of ctDNA increases substantially for tumors with overall volume greater than 65 mL. Possible biological mechanisms underlying such a finding could include the presence of larger areas of tumor necrosis and/or greater amounts of apoptosis leading to greater ctDNA shedding or possibly that degradation of ctDNA becomes saturated or less efficient with larger amounts in circulation leading to increased likelihood of mutation detection.
The secondary endpoint of OS yielded interesting results as well. Multivariate analysis showed that age, mutation detection and presence of bone metastases were independently associated with OS, but there was also a trend toward significance for total tumor volume greater than 65 mL and presence of liver metastases. Much work has been done on defining the oligometastatic disease state which has been defined variably, e.g., as a maximum of 3 or 5 metastatic sites. It is reasonable to hypothesize that total tumor volume as a continuous, absolute quantity may provide a logical and biologically supported means by which to better stratify prognosis in patients with oligo- and polymetastatic NSCLC than by number of metastases alone.
There are several features of this analysis that distinguish it from prior studies. First, ascertainment of tumor volume in the present study was performed through careful manual examination of axial images and delineation of tumor on each image. Other studies have estimated tumor volume based on the longest lesion diameter (15,16) or three-dimensional measurements on axial imaging (13,14). One of these studies specifically addressed patients with NSCLC (14), and in that case, half of the patients with stage IV cancer were not considered by the authors to have a reliably assessable tumor volume. Our method allowed us to assess tumor volume in all cases of stage IV cancer that were encountered. Second, the large size of our patient cohort and the granularity of our data allowed us to explore the relationship between tumor volume and mutation detection to a greater degree than has been previously done. Third, we found that total tumor volume is an independent predictor of OS, even when accounting for factors such as detection of mutations in cfDNA. This risk factor could be valuable in stratifying patients with metastatic and locally advanced disease.
This study has limitations. Mutation detection is an imperfect surrogate measure for assessment of total ctDNA shedding. This study used an assay that can detect alterations in 21 genes, which does not capture a full representation of alterations in cfDNA. The present study found mutations in 71% of patients, and our group previously published a larger cohort using the same NGS panel with a detection rate of 64% (3). Other recent studies published using larger gene panels have found higher rates of mutation detection, such as a 36-gene panel that found mutations in 77% of patients (30), and a 62-gene panel that found evidence of ctDNA in 80% of patients (31). Such larger panels may thus have greater mutation detection sensitivity. On the other hand, by focusing on high-yield genetic alterations in NSCLC, the present analysis remains a clinically relevant investigation. Newer NGS assays are developing increasing analytical sensitivity in addition to larger panel size, so while our data may indicate a greater clinical relevance for NGS assays performed on larger tumors, limitations in smaller tumors and earlier stage disease may diminish with coming technological advances. More sensitive panels may well detect mutations in smaller tumors, which would reduce the threshold of tumor volume identified in this study. Further study will be needed to evaluate a tumor volume threshold for other assays to further characterize tumor volume as a predictor of mutation detection and determine what degree of variability exists.
Clonal hematopoiesis (CH) was also not controlled for in this assay, so it is possible that some detected mutations may be attributable to CH instead of the patient’s cancer. Another consideration is that all mutations may not be shed equivalently, perhaps due to copy number alterations and other factors. For the purposes of this analysis, it was necessary to assume that all mutations are shed equally; while we are unaware of any published data to illuminate this area, future work may find differences in shedding between mutations. A limitation in the real-world application of our methods is that volume ascertainment as performed in this study is time intensive, and as a result, tumor volume delineation was only checked by a second observer in select cases. However, future investigation may be done to automate the collection of detailed volumetric data to reduce the difficulty in obtaining such information and improve clinical applicability. Lastly, these results may not be generalizable outside the setting of NSCLC.
Conclusions
In conclusion, we found that total tumor volume greater than 65 mL was a significant predictor of cfDNA mutation detection as assessed by an NGS panel assay in a group of patients with advanced NSCLC. Total tumor volume greater than 65 mL trended toward a significant association with OS, raising the possibility that tumor volume may be an important risk factor in prognostication of metastatic NSCLC. More research is warranted to investigate tumor volume as a predictor of cfDNA mutation detection and as a prognostic risk factor in lung cancer and other primary disease sites.
Acknowledgments
The Molecular Diagnostics Service in the Department of Pathology at Memorial Sloan Kettering Cancer Center, the Marie-Josee and Henry R. Kravis Center for Molecular Oncology.
Funding: This research was partially supported by the NIH/NCI Cancer Center Support Grant/Core Grant (P30 CA008748). No sponsor participated in study design, data analysis or interpretation, writing the report, or the decision to submit the article for publication. The funding source had no involvement in the in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-164/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-164/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-164/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-164/coif). MO receives honorarium from Jazz Pharmaceuticals, Novartis, PharmaMar, Targeted Oncology and OncLive. AR performs consultancy and advisory roles with AstraZeneca. LPL and ML own stock in Resolution Bioscience. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Institutional Review Board of Memorial Sloan Kettering Cancer Center (Protocol 12-245) and informed consent was taken from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33. [Crossref] [PubMed]
- Kapeleris J, Ebrahimi Warkiani M, Kulasinghe A, et al. Clinical Applications of Circulating Tumour Cells and Circulating Tumour DNA in Non-Small Cell Lung Cancer-An Update. Front Oncol 2022;12:859152. [Crossref] [PubMed]
- Sabari JK, Offin M, Stephens D, et al. A Prospective Study of Circulating Tumor DNA to Guide Matched Targeted Therapy in Lung Cancers. J Natl Cancer Inst 2019;111:575-83. [Crossref] [PubMed]
- Razavi P, Li BT, Brown DN, et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med 2019;25:1928-37. [Crossref] [PubMed]
- Stroun M, Anker P, Maurice P, et al. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology 1989;46:318-22. [Crossref] [PubMed]
- Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223-38. [Crossref] [PubMed]
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426-37. [Crossref] [PubMed]
- Heitzer E, Haque IS, Roberts CES, et al. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet 2019;20:71-88. [Crossref] [PubMed]
- Diehl F, Li M, Dressman D, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A 2005;102:16368-73. [Crossref] [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [Crossref] [PubMed]
- Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018;359:926-30. [Crossref] [PubMed]
- Avanzini S, Kurtz DM, Chabon JJ, et al. A mathematical model of ctDNA shedding predicts tumor detection size. Sci Adv 2020;6:eabc4308. [Crossref] [PubMed]
- Valpione S, Gremel G, Mundra P, et al. Plasma total cell-free DNA (cfDNA) is a surrogate biomarker for tumour burden and a prognostic biomarker for survival in metastatic melanoma patients. Eur J Cancer 2018;88:1-9. [Crossref] [PubMed]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [Crossref] [PubMed]
- Ascierto PA, Minor D, Ribas A, et al. Phase II trial (BREAK-2) of the BRAF inhibitor dabrafenib (GSK2118436) in patients with metastatic melanoma. J Clin Oncol 2013;31:3205-11. [Crossref] [PubMed]
- McEvoy AC, Warburton L, Al-Ogaili Z, et al. Correlation between circulating tumour DNA and metabolic tumour burden in metastatic melanoma patients. BMC Cancer 2018;18:726. [Crossref] [PubMed]
- Khier S, Lohan L. Kinetics of circulating cell-free DNA for biomedical applications: critical appraisal of the literature. Future Sci OA 2018;4:FSO295. [Crossref] [PubMed]
- Wong SQ, Raleigh JM, Callahan J, et al. Circulating Tumor DNA Analysis and Functional Imaging Provide Complementary Approaches for Comprehensive Disease Monitoring in Metastatic Melanoma. JCO Precis Oncol 2017;1:1-14. [Crossref] [PubMed]
- De Mattos-Arruda L, Mayor R, Ng CKY, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun 2015;6:8839. [Crossref] [PubMed]
- Morbelli S, Alama A, Ferrarazzo G, et al. Circulating Tumor DNA Reflects Tumor Metabolism Rather Than Tumor Burden in Chemotherapy-Naive Patients with Advanced Non-Small Cell Lung Cancer: 18F-FDG PET/CT Study. J Nucl Med 2017;58:1764-9. [Crossref] [PubMed]
- Winther-Larsen A, Demuth C, Fledelius J, et al. Correlation between circulating mutant DNA and metabolic tumour burden in advanced non-small cell lung cancer patients. Br J Cancer 2017;117:704-9. [Crossref] [PubMed]
- Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446-51. [Crossref] [PubMed]
- Bratman SV, Yang SYC, Iafolla MAJ, et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat Cancer 2020;1:873-81. [Crossref] [PubMed]
- Tie J, Wang Y, Cohen J, et al. Circulating tumor DNA dynamics and recurrence risk in patients undergoing curative intent resection of colorectal cancer liver metastases: A prospective cohort study. PLoS Med 2021;18:e1003620. [Crossref] [PubMed]
- Garcia-Murillas I, Chopra N, Comino-Méndez I, et al. Assessment of Molecular Relapse Detection in Early-Stage Breast Cancer. JAMA Oncol 2019;5:1473-8. [Crossref] [PubMed]
- Lo YM, Chan AT, Chan LY, et al. Molecular prognostication of nasopharyngeal carcinoma by quantitative analysis of circulating Epstein-Barr virus DNA. Cancer Res 2000;60:6878-81. [PubMed]
- Lin JC, Wang WY, Chen KY, et al. Quantification of plasma Epstein-Barr virus DNA in patients with advanced nasopharyngeal carcinoma. N Engl J Med 2004;350:2461-70. [Crossref] [PubMed]
- Paweletz CP, Sacher AG, Raymond CK, et al. Bias-Corrected Targeted Next-Generation Sequencing for Rapid, Multiplexed Detection of Actionable Alterations in Cell-Free DNA from Advanced Lung Cancer Patients. Clin Cancer Res 2016;22:915-22. [Crossref] [PubMed]
- Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 2015;17:251-64. [Crossref] [PubMed]
- Remon J, Lacroix L, Jovelet C, et al. Real-World Utility of an Amplicon-Based Next-Generation Sequencing Liquid Biopsy for Broad Molecular Profiling in Patients With Advanced Non-Small-Cell Lung Cancer. JCO Precis Oncol 2019;3. ePO. [Crossref] [PubMed]
- Schrock AB, Welsh A, Chung JH, et al. Hybrid Capture-Based Genomic Profiling of Circulating Tumor DNA from Patients with Advanced Non-Small Cell Lung Cancer. J Thorac Oncol 2019;14:255-64. [Crossref] [PubMed]


