
BRAF RNA is prognostic and widely expressed in lung adenocarcinoma
Introduction
Molecular profiling identified 30–50% of non-small cell lung cancer (NSCLC) patients with a targetable genomic alteration (1). The majority harbor EGFR and KRAS mutations (2-5). However, ALK, ROS1, RET rearrangement, and BRAF, ERBB2, MET, and PIK3CA mutations were identified in only 0.8–6.0%, and these low-frequency targets translated into a therapeutic aspect for a limited number of patients (4,6-8). Gain-of-function mutations in the BRAF gene are around 8% of all human malignancies. About 50% of melanoma patients harbor V600E mutations that are successfully treated with various BRAF inhibitors (9).
In contrast to melanoma, BRAF mutations are identified in 2–4% of lung adenocarcinomas (LADCs) (10-12). The BRAF gene encodes RAF serine/threonine kinase proteins, including ARAF, BRAF, and CRAF isoforms. RAF mutation is associated with activating the mitogen-activated protein kinase (MAPK) pathway promoting cell growth, proliferation, and survival (13). Overexpression of BRAF constitutively results in an increase in oncogenic signaling through MEK.
Following BRAF inhibition, epigenetic reprogramming might drive changes in differentiation status and establish novel gene expression programs that diminish the cellular demand for BRAF or MAPK signaling. This mechanism, known as phenotypic- or epigenetic switching, produces fractional response even in genetically homogeneous populations of tumor cells. However, they appear to be controlled by lineage-dependent, epigenetic processes that constantly change the state of susceptibility in tumor cells (14,15). Nevertheless, aside from genetic alterations, epigenetic plasticity might impact a tumor cell’s BRAF reliance and responsiveness to treatment.
While most studies focus on the low-frequency genomic landscape of BRAF, the RNA expression patterns of the wild-type (WT) BRAF gene expression have not been investigated in NSCLC. This study performed a transcriptomic analysis of LADC with RNAscope in situ hybridization (ISH) assay, which significantly advances target-specific signal detection. We aimed to analyze the associations between BRAF expression, clinicopathological parameters, and outcomes of 64 LADC patients using RNAscope. We present the following article in accordance with the REMARK reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-449/rc).
Methods
Ethical statement
We performed the study based on the Helsinki Declaration of the World Medical Association study guidelines (as revised in 2013), and the Hungarian Scientific and Research Ethics Committee of the Medical Research Council approved the study (No. 2307-3/2020/EÜIG). All the patients provided informed consent. After the clinical and pathological data were collected, patient identifiers were removed; therefore, individual patients cannot be identified directly or indirectly.
Study population
A total of 64 histologically confirmed LADC patients, with available primary tumor tissue were included in our study. All patients underwent surgical resection or lung biopsy (n=37 Stage I–II, n=17 Stage III, and n=10 Stage IV) from 2006 to 2013 at the National Koranyi Institute of Pulmonology. Formalin-fixed, paraffin-embedded (FFPE) tissue samples from primary tumors were obtained at lung resection surgery for Stage I–II patients (n=37) and with lung biopsy for Stage III–IV (n=27) patients. An expert pathologist determined pathological characteristics. Clinicopathological characteristics of patients, including age, gender, stage, comorbidities [diabetes, chronic obstructive pulmonary disease (COPD)], blood work at diagnosis and tumor pathology [tumor necrosis, tumor grade, vascular involvement, peritumoral infiltration, and programmed death ligand 1 (PD-L1) status] and treatments are described in Table 1. Patients were treated according to the current international guidelines with the standard of care therapy. Platinum-based doublet therapy were administered as a frequent oncotherapy. Patient flow diagram shows cohort characteristics and patient selection in Figure S1.
Table 1
| Parameter | Group A | Group B | Missing values |
|---|---|---|---|
| Age (years) | <60 [52% (n=33)] | ≥60 [48% (n=31)] | 0 |
| Sex | Male [53% (n=34)] | Female [47% (n=30)] | 0 |
| Smoking | Never [10% (n=5)] | Smoker [90% (n=44)] | 23% (n=15) |
| Stage (I–II vs. III–IV ) | I–II [58% (n=37)] | III–IV [42% (n=27)] | 0 |
| Diabetes | No [91% (n=58)] | Yes [9% (n=6)] | 0 |
| COPD | No [74% (n=45)] | Yes [26% (n=16)] | 5% (n=3) |
| Tumor grade (2 vs. 3) | Grade 2 [72% (n=46)] | Grade 3 [28% (n=18)] | 0 |
| Tumor necrosis | No [20% (n=13)] | Yes [80% (n=51)] | 0 |
| Vascular involvement | No [69% (n=44)] | Yes [31% (n=20)] | 0 |
| Peritumoral infiltration | Moderate 1–2 [73% (n=47)] | High 3 [27% (n=17)] | 0 |
| PD-L1 immune cells | Low <1.5 [23% (n=9)] | High ≥1.5 [77% (n=30)] | 39% (n=25) |
| PD-L1 tumor cells | Low <1.5 [74% (n=28)] | High ≥1.5 [26% (n=10)] | 40% (n=26) |
| PD-1 immune cells | Low <1.5 [46% (n=18)] | High ≥1.5 [54% (n=21)] | 39% (n=25) |
| BRAF expression | Low <2 [36% (n=17)] | High ≥2 [64% (n=47)] | 0 |
| Sample acquisition | Resection [58% (n=37)] | Biopsy [42% (n=27)] | 0 |
| Adjuvant chemotherapy | No [60% (n=38)] | Yes [40% (n=26)] | 0 |
| IIIB–IV chemotherapy | No [63% (n=40)] | Yes [37% (n=24)] | 0 |
Group A, Low/No clinicopathological features; Group B, High/Yes clinicopathological features. COPD, chronic obstructive pulmonary disease; PD-L1, programmed death ligand 1; PD-1, programmed death 1.
Tissue processing
Tissue samples were fixed and processed into paraffin blocks. Tissue microarrays (TMA) were constructed from FFPE blocks as previously described (16,17). Briefly, 4-micron sections from each tissue block were prepared using an HM-315 microtome (Microm, Boise, ID, USA) and placed on charged glass slides (Colorfrost Plus, #22-230-890; Fisher, Racine, WI, USA). Slides were stained for H&E on an automated Tissue-Tek Prisma staining platform (Sukura, Osaka, Japan). A laboratory pathologist reviewed H&E slides. Marked and stained sections were used to guide the technician to the location for core tissue removal. Three 3-mm cores of tissue were taken from each donor tissue block for primary tumors and seated into a recipient paraffin block in a positionally encoded array format (MP10 3.0 mm tissue punch on a manual TMA instrument; Beecher Instruments, Sun Prairie, WI, USA).
RNAscope
RNAScope® ISH assay was performed on TMAs from human adenocarcinomas using RNAScope® Multiplex Fluorescent Kit v2 according to the manufacturer’s instructions (Advanced Cell Diagnostics Pharma Assay Services, Newark, CA, USA). Briefly, 4 µm formalin-fixed paraffin-embedded TMA sections were pretreated with antigen-retrieval buffer, heat, and protease prior to hybridization with the following target oligo probes: 3plex-Hs-Positive Control Probe (ACDBio, cat: 320861), 3plex-Hs-Negative Control Probe (ACDBio, cat: 320871), Hs-BRAF-C1 (ACDBio, cat: 595251). Preamplifier, amplifier, and AMP-labeled oligo probes were then hybridized sequentially, followed by fluorogenic precipitate development. Cy3 (red) fluorochrome was used to visualize binding spots for amplified probes. Each sample was quality controlled for RNA integrity with a positive control probe specific to housekeeping genes, while a negative control probe set was used to assess background fluorescence. The pretreatment conditions were optimized to establish the maximum signal-to-noise ratio. Specific RNA staining signal was identified as red punctate dots. Nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI) appearing light blue. Imaging was performed with Zeiss LSM 780 Confocal microscope. We performed RNAScope® - IHC codetection, anti-E-cadherin monoclonal antibody (Cell Signaling, cat: #3195) to identify tumor cells of epithelial origin. The primary antibody was applied overnight before the hybridization steps. Antigen-antibody complexes were crosslinked by neutral buffered formalin for 30 minutes before protease treatment. Slides were incubated with ALEXA A488 (green) anti-mouse secondary antibody for 45 minutes after signal amplification and detection of RNAScope® probes.
Scoring with QPath Software
For the quantitative evaluation of RNA expression, the Qpath software package was used (18). After selecting representative regions on slides containing no positive RNAscope® staining, we measured the total intensity of the selected background regions and calculated the average background intensity (Average Intensity of Background per Pixel) using the following equation:
To quantify the average intensity per single dot, first, we selected at least 20 single signal dots for every visual field and measured each dot’s area and total intensity. Then, we used the area of each dot to screen whether the dot is a true single dot and calculated the average intensities for every single dot:
To measure the total area of the region of interest (ROI) and total intensity of ROI, we used average intensity per single dot. We calculated the total dot number in the ROI:
Next, we counted the number of cells in the ROI by counting DAPI positive nuclei and used this value to calculate the average dot number per cell.
We used the DAPI nuclear staining to define each cell region by assigning the cell’s radius and assigning each cell as one ROI. Then, we counted the dot number in each ROI as previously described. We calculated an integrated expression score [0–3] for all tumor cores based on dots’ average density and raw intensity data, where k-means clustering method was used to determine cut-offs. We included three cores per patient in all TMAs, and an average BRAF expression score was calculated for every patient. All intensity measurements were carried out by an LSM780 Zeiss confocal microscope and Zen Blue software package.
Immunohistochemistry, histopathology, and scoring
Four-µm sections were cut from every FFPE TMA block for IHC staining. Staining was performed as previously described (16,17) on a Leica Bond RX autostainer using rabbit monoclonal antibody for PD-L1 diluted 1:300 (CST, cat: 13684S), rabbit monoclonal antibody for programmed death 1 (PD-1) diluted 1:400 (CST, cat: 86163). Briefly, slides were stained using the Bond Polymer Refine Detection kit (#DS9800) with Leica IHC Protocol F, and epitope retrieval was performed at low pH for twenty minutes. Clearing and dehydration of slides were automated on a Tissue-Tek Prisma platform and then coverslipped using a Tissue-Tek Film coverslipper. Counterstaining was performed with hematoxylin. For tumor-cell-PD-L1 expression, the Allred score was calculated based on staining intensity [0–3]. The number of positive cells was calculated for each TMA core to assess immune cell-PD-L1 and immune cell-PD-1 expression. Immune cell PD-L1- and PD-1-positive immune cell counts were normalized to a 4-level scale [0–3] based on the k-means clustering method. Normalized cell numbers and determined Allred scores of the three cores were averaged for every patient. Tumor cells and immune cells were identified by routine HE staining. Staining protocols were optimized on healthy human lung and tonsil tissues.
Datasets
IHC staining images of BRAF protein expression in LADC tissues (n=10) were analyzed from the Human Protein Atlas (HPA) database [http://www.proteinatlas.org/, (19)]. All datasets were obtained from the literature, and the ethics declarations indicated that all patients supplied written informed consent. RNA-seq data are reported as average fragments per kilobase of exon per million reads mapped (FPKM) generated by The Cancer Genome Atlas (TCGA) accessed from the HPA database (n=500). Based on the FPKM value of each gene retrieved from TCGA data, patients were classified into two expression groups and the correlation between expression level and patient survival was examined. The prognosis of each group of patients was examined by Kaplan-Meier survival estimators, and the survival outcomes of the two groups were compared by log-rank tests.
Statistical analyses
We utilized the Shapiro-Wilk test to determine the appropriate statistical methods for the dataset. We used non-parametric tests Mann-Whitney U-test, to compare BRAF RNA expression between patient groups according to clinicopathological characteristics and PD-L1 expression. Spearman’s correlation coefficient was calculated to evaluate clinicopathological parameters’ statistical correlation. P values <0.05 indicate the significance, and all P values were two-sided. Survival analysis was performed using Kaplan-Meier curves and Cox proportional hazard regression, and we compared the survival curves with the log-rank test. The backward regression method was used for multivariate Cox regression. MedCalc Software package was applied for survival analyses. Prism software was used for statistical tests and data visualisation.
Results
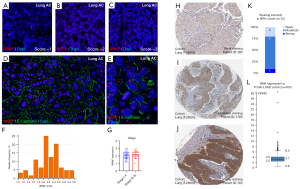
Tumor samples of 64 LADC patients were included and stained by RNAscope in our study, where an average BRAF RNA expression score was identified for each individual patient (Figure S1). We found no patients with a score below 1, meaning that BRAF is expressed at minimum baseline levels in LADCs. Our cohort’s average BRAF expression score was 2.17±0.46, with an overall low level of intratumoral heterogeneity, where expression scores across TMA cores showed a strong positive correlation (average r=0.62, P<0.001). Figure 1A-1C show representative RNAscope stainings on samples scored between 1 and 3. BRAF RNA was predominantly expressed inside E-cadherin-positive cancer cells with only scattered expression in the stroma (Figure 1D,1E). BRAF expression showed no correlation with our stratified stage score (Figure S2, r=0.195, P=0.127). Figure 1F shows the relative distribution frequency of aggregate scores given to patients regarding BRAF expression. There was no significant difference in BRAF expression between early-stage (I–II) and advanced-stage (III–IV) patients (2.16 vs. 2.16, P=0.317, Figure 1G). Protein expression of the BRAF gene was validated by accessing a public dataset from the HPA. Representative TMA samples show perinuclear and cytoplasmic expression patterns of BRAF protein stained with IHC in the HPA LADC cohort. Similar to the RNAscope staining pattern, IHC also shows that tumor nests are diffusely stained, whereas BRAF-positive cells occur only scattered in the stroma (Figure 1H-1J). In the HPA cohort (n=10), the distribution of protein expression (Figure 1K) roughly follows the RNA expression pattern evaluated by RNAscope (Figure 1F). Box plot shows the median and 25th and 75th percentile values of relative RNA expression (FPKM) in the TCGA-LUAD cohort (Figure 1L).
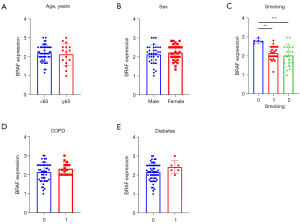
Next, we analyzed the associations between clinicopathological characteristics and BRAF expression scores. There was no significant difference according to BRAF expression and age (≥60 vs. <60 years, 2.29 vs. 2.16, P=0.916, Figure 2A), and sex [2.16 (male) vs. 2.2 (female) P=0.46, Figure 2B]. Smoking, however, showed a significant negative correlation with BRAF expression (r=−0.29, P=0.043), and never smokers express significantly higher levels of BRAF RNA compared to current smokers (2.83 vs. 2.12, P=0.004), and ex-smokers (2.83 vs. 2.17, P<0.001, Figure 2C). In contrast, the presence of comorbidity of lung parenchyma (COPD vs. non-COPD; 2.25 vs. 2.16, P=0.295, Figure 2D) or diabetes [2.37 (present) vs. 2.16 (not present), P=0.185, Figure 2E] showed no significant difference concerning BRAF expression. Correlation coefficients (Spearman’s) and P values for every parameter are shown in Tables S1,S2.
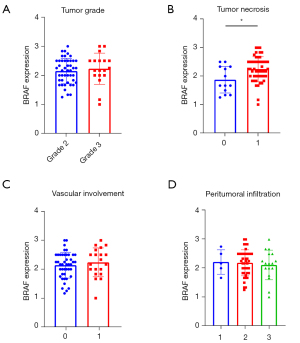
An expert pathologist assessed the presence of tumor necrosis and vascular involvement on tissue samples. Histopathological characterization included peritumoral scoring infiltration [0–3] and tumor grade [1–3] with routine HE-staining. According to peritumoral infiltration, all tumors had a score between 1 and 3. Tumor necrosis showed a significant positive correlation with clinical stage (r=0.35, P=0.005) and tumor grade (r=0.32, P=0.009), and there was a significant negative correlation with smoking (r=−0.34, P=0.018) (Figure S2). Furthermore, tumor necrosis showed a significant positive correlation with BRAF expression (r=0.3, P=0.014) (Figure S2). BRAF expression was not significantly different in grade 3 vs. grade 2 tumors (2.2 vs. 2.16, P=0.449, Figure 3A), however patients with tumor necrosis were detected to express significantly higher levels of BRAF RNA (2.25 vs. 1.66, P=0.014, Figure 3B). The presence of vascular involvement (2.25 vs. 2.16, P=0.427, Figure 3C), or peritumoral infiltration (P=0.9, Figure 3D) showed no significant associations with BRAF expression. Necrosis exhibited no correlation with tumor cell PD-L1 expression (rs =0.288, P=0.071), but a significant positive correlation with tumor grade (rs =0.322, P=0.01) and we found a significant negative correlation with smoking (rs =0.338, P=0.02). Tumor grade, vascular involvement, and peritumoral infiltration were not correlated significantly with BRAF expression (Figure S2).
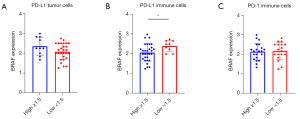
Next, we compared BRAF expression according to the PD-L1 phenotype; we compared high PD-L1 expressor (aggregate score, ≥1.5) patients with low expressors (aggregate score, 0–1.5). PD-L1 expression was assessed on tumor cells (average score, 1.01±0.91) and immune cells (average score, 1.93±0.62). PD-1 expression was evaluated on immune cells (average score, 1.53±0.64). There was no significant difference in BRAF RNA expression between tumor PD-L1-high vs. low patients (2.25 vs. 2.08, P=0.124, Figure 4A). In contrast, BRAF expression showed a significant difference between immune cell PD-L1-high vs. low patients (2.08 vs. 2.5, P=0.03, Figure 4B), but no significant difference was identified between immune cell PD-1-high- and PD-1-low patients (2.2 vs. 2.16, P=0.721, Figure 4C).
Next, we analyzed the association between BRAF RNA expression and preoperative blood work parameters, including an absolute number of leukocytes, lymphocytes, neutrophils, and platelets along with neutrophil to lymphocyte ratio (NLR) and platelet to lymphocytes ratio (PLR). We found no significant correlations between blood work parameters and BRAF expression (Figure S2). We also compared patients with BRAF-high (2.0–3.01) and -low expression (0–1.99) and found no significant differences in any comparison with any blood work parameter (Figure S3). Thrombocyte number showed a significant negative correlation with smoking (r=0.29, P=0.44), numbers of leukocytes (r=0.25, P=0.046) and lymphocytes (r=0.27, P=0.033) and a significant positive correlation with clinical stage and lymphocyte number with female sex (r=0.25, P=0.044) (Figure S2).
To reveal the association of clinical outcomes with WT BRAF expression, we performed a Kaplan-Meier survival analysis stratifying patients to BRAF-high (2.0–3.0) and BRAF-low (1.0–1.99) expressors. Survival was not significantly different between early- and late-stage patients (Figure S4), and Chi-squared test showed no significant association between BRAF-high and -low phenotype and early- or late-stage disease (P=0.124). Patients with low BRAF expression scores showed significantly increased OS, compared to BRAF-high patients [mean OS: 61±30 vs. 28±13 months, hazard ratio (HR): 0.259, 95% CI: 0.09521 to 0.7043, P=0.0081, Log-rank test, Figure 5A]. This was supported by Spearman correlation, which showed a significant negative correlation between BRAF expression and OS (r=−0.41, P<0.001, Figure S2). Figure 5B shows diverging Kaplan-Meier curves for overall survival (OS), according to patient quadrants regarding BRAF expression (mean OS, 54±34 vs. 64±29 vs. 34±20 vs. 28±13 months, P=0.002, Log-rank test for trend).
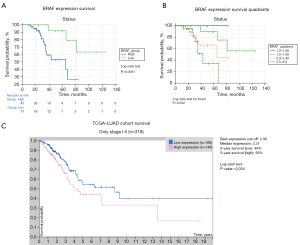
While tumor necrosis showed a significant negative correlation with OS (r=−0.28, P=0.023, Figure S2), survival analysis showed no significant decrease in survival (82±10 vs. 78±9 months, P=0.492). Additional Kaplan-Meier curves show OS according to sex (P=0.833), stage (P=0.668), diabetes (P=0.037), COPD (P=0.036), tumor necrosis (P=0.492), peritumoral infiltration (P=0.041) and vascular involvement (P=0.038) in Figure S4. To confirm our findings in publicly available datasets, the TCGA database was accessed and Kaplan-Meier curves were generated with optimized cut-off values generated via the HPA platform. While BRAF high expressor (cut-off: 3.38 FPKM) LADC patients exhibited significantly decreased OS compared to BRAF low expressors (5-year survival: 44% vs. 56%, P=0.034, n=318, Figure 5C) in early stage (I–II) disease, there was no significant difference in survival between BRAF high- vs. low expressors (cut-off: 2.41 FPKM), when pooling early- and late-stage LADC patients (P=0.25, n=500, not shown).
BRAF expression was identified as a significant negative prognostic factor (P=0.01) according to univariate Cox proportional hazard regression. In our analysis, the presence of COPD (P=0.029), diabetes (P=0.044), and vascular involvement (P=0.046) were also significant predictors of OS. According to univariate analysis (Table 2), the level of peritumoral infiltration was borderline significant (0.058). Next, we included significant parameters in our multivariate model, where we used backward elimination to perform the regression (Table 3). Parameters with p>0.1 were eliminated (COPD, vascular involvement). Apart from the BRAF score (P=0.025), peritumoral infiltration was an independent predictor of OS (P=0.022). Harral’s C-index for the goodness of the model was 0.72 (95% CI: 0.630 to 0.811).
Table 2
| Covariate | B | SE | Wald | P value | Exp (B) | 95% CI of Exp (B) |
|---|---|---|---|---|---|---|
| Age | 0.02886 | 0.03201 | 0.8124 | 0.3674 | 1.0293 | 0.9667 to 1.0959 |
| Sex (male vs. female) | 0.1026 | 0.4882 | 0.04412 | 0.8336 | 1.1080 | 0.4255 to 2.8850 |
| Clinical stage | 0.06772 | 0.08934 | 0.5746 | 0.4484 | 1.0701 | 0.8982 to 1.2748 |
| Smoking | 0.2628 | 0.4571 | 0.3306 | 0.5653 | 1.3006 | 0.5333 to 3.1716 |
| BRAF score | 1.5580 | 0.6064 | 6.6015 | 0.0102* | 4.7495 | 1.4470 to 15.5890 |
| COPD (0–1) | 1.1299 | 0.5157 | 4.8005 | 0.0285* | 3.0953 | 1.1265 to 8.5051 |
| Diabetes (0–1) | 1.3789 | 0.6833 | 4.0731 | 0.0436* | 3.9707 | 1.0406 to 15.1518 |
| PD-L1 tumor cells | 0.8042 | 0.4944 | 2.6463 | 0.1038 | 0.4474 | 0.1698 to 1.1791 |
| PD-L1 immune | −0.1492 | 0.4574 | 0.1063 | 0.7443 | 0.8614 | 0.3514 to 2.1115 |
| PD-1 immune | 0.04450 | 0.3203 | 0.01930 | 0.8895 | 1.0455 | 0.5599 to 1.9523 |
| Tumor necrosis (0–1) | −0.3924 | 0.5762 | 0.4637 | 0.4959 | 0.6754 | 0.2183 to 2.0896 |
| Tumor grade (2 vs. 3) | 0.1084 | 0.5346 | 0.04115 | 0.8393 | 1.1145 | 0.3908 to 3.1783 |
| Vascular involvement | 0.9697 | 0.4868 | 3.9675 | 0.0464* | 2.6372 | 1.0156 to 6.8479 |
| Peritumoral infiltration | −0.8301 | 0.4371 | 3.6070 | 0.0575* | 0.4360 | 0.1851 to 1.0269 |
*, significant predictors. B, coefficient for the constant; SE, standard error around the coefficient; Wald, Wald chi-square test; Exp (B), exponentiation of the B coefficient (odds ratio); COPD, chronic obstructive pulmonary disease; PD-L1, programmed death ligand 1; PD-1, programmed death 1.
Table 3
| Covariate | B | SE | Wald | P value | Exp (B) | 95% CI of Exp (B) |
|---|---|---|---|---|---|---|
| BRAF score | 1.4945 | 0.6655 | 5.0432 | 0.0247* | 4.4570 | 1.2094 to 16.4250 |
| Diabetes | 1.2292 | 0.7450 | 2.7220 | 0.0990 | 3.4185 | 0.7937 to 14.7241 |
| Peritumoral infiltration | −1.0708 | 0.4668 | 5.2621 | 0.0218* | 0.3427 | 0.1373 to 0.8557 |
*, significant predictors. B, coefficient for the constant; SE, standard error around the coefficient; Wald, Wald chi-square test; Exp (B), exponentiation of the B coefficient (odds ratio).
Discussion
Overexpression of tumor intracellular proliferation pathways is associated with drug resistance and disease progression. Distinct targetable genetic driver alterations have been explored in LADC; despite BRAF being a critical and targetable signaling molecule, mutations are present in only 2–4% of NSCLC cases.
The activation of the MAPK pathway promotes cell growth, proliferation, and survival. Cardarella et al. reported that 56.7% of BRAF mutations were the activating V600E (20,21) whereas the rest of BRAF mutations appear to be the inactivating type in lung cancer (22,23). Although some reports have correlated BRAF mutation in NSCLC with a poorer outcome and reduced efficacy of platinum doublets, the prognostic implication of BRAF V600E mutated NSCLC remains unclear (24,25). A recent study showed that BRAF-mutated NSCLC patients treated with standard of care therapy have an inferior prognosis (25). In contrast, higher susceptibility to immune checkpoint inhibitors (ICIs) was associated with mutated BRAF status (25).
The expression patterns of the WT BRAF RNA have not been investigated in NSCLC that have potential translational relevance through enhanced downstream signalization (26,27). Moreover, we used in situ transcriptomic analysis, which is a quick and cost-effective methodology to identify patient subsets with BRAF spatial distribution. We also analyzed associations with clinical outcomes. Since ICI is implemented in the frontline and subsequent therapeutic lines, we also characterized our cohort for PD-L1 expression, a key biomarker in NSCLC. Accordingly, we explored the potential relationships of PD-L1 relative to BRAF RNA expression, which might help plan future clinical studies. We found that BRAF expression was negatively correlated with PD-L1 expression in immune cells but showed no significant association with PD-L1 expression in tumor cells or PD-1 expression. ISH showed that BRAF RNA was widely expressed in tumor nests, inside E-cadherin-positive cancer cells only with scattered expression in the stroma. While sex, age, and clinical stage showed no association with BRAF expression, never smokers express significantly higher BRAF RNA levels than current smokers and ex-smokers. There were no differences in tumor grade, vascular involvement, and peritumoral infiltration according to BRAF RNA expression. Notably, BRAF RNA was expressed in the early and late stages of the disease. Therefore our results showed that BRAF is widely expressed in LADC and is possibly advantageous for clinical studies that might need no preselection for patient recruitment. Additionally, our data is in line with others that showed a no different distribution of BRAF mutations across major clinicopathological characteristics; however, others used genomic sequencing compared to our RNA scope expression analysis (25,28).
Our data showed that necrotic tumors tend to show intense BRAF RNA expression. An increased level of BRAF RNA expression was an independent adverse prognostic factor, whereas the extent of peritumoral infiltration was an independent favorable prognostic factor according to our multivariate analysis. The negative prognostic role of BRAF RNA expression was also confirmed in an independent TCGA cohort of LADC patients (n=318), but only in the case of early stage (I–II) disease. the Necrosis was correlated with tumor cell PD-L1 expression, tumor grade, and smoking. Previously, others reported an association between tumor necrosis and tumor cell PD-L1 expression (29), whereas in our cohort, tumor necrosis correlated with stage, tumor grade and smoking.
BRAF kinase has an essential role in intracellular signaling, facilitating signal transduction from membrane receptors to the nucleus following epithelial growth factor receptor (EGFR) activation (30). The co-inhibition of MEK- and BRAF kinases have improved outcomes in some BRAF-mutated malignities; however, most cases still develop some form of resistance (31-33). While BRAF kinase is normally deactivated in healthy tissues through a negative feedback loop, mutations in the BRAF gene result in persistent activation of downstream cell signaling in the MAPK pathway, leading to uncontrolled cell growth and proliferation (34-36). Others showed in Caucasian lung cancer patient cohorts that smoking status was associated with a non-V600E mutation (20,37,38). In contrast, a study on an East Asian LADC cohort and a meta-analysis of 16 studies found the V600E subtype more common in non-smokers. The same studies showed associations with decreased chemosensitivity and worse prognosis (39,40). This is in line with our results, where increased BRAF RNA expression was also associated with non-smoker patient history and detrimental OS. In our cohort, most patients showed strong diffuse BRAF RNA signals that cannot be explained by potential V600E mutations since its frequency hardly reaches 3–5% in LADC patients. This might mean that WT BRAF can still be overexpressed in the absence of mutation in the BRAF gene due to alternative intracellular signaling pathways or the tumor microenvironment. This is underlined by the fact that the gene’s protein product is also detectable by IHC with at least moderate staining intensity in 80% of samples, according to the HPA database. Activation of the BRAF-MAPK pathway is always preceded by the interaction between the guanosine-nucleotide-binding protein RAS and a Receptor Tyrosine Kinase (RTK). Because BRAF selectively binds to active RAS (41), hypothetically, increased RAS activation and RTK function might have a positive upstream effect on BRAF signaling, even in the absence of activating mutations.
Others reported a retrospective multi-centre chart review that assessed the response of BRAF positive NSCLC to immune checkpoint inhibitors. The authors assessed PD-L1 status, tumor mutational burden (TMB) and microsatellite instability (MSI) in 39 patients. BRAF-mutated NSCLC was more likely to have high expression of PD-L1 (42). A case study with five patients reported similar findings (43). In contrast, we found that PD-L1 expression in tumor cells is independent from the extent of BRAF RNA expression, and immune cell PD-L1 status negatively correlates with BRAF RNA quantity. The discrepancy can be explained by the different patient populations and the relatively high number of potential inactivating non-V600E mutations in the cited cohorts. In a retrospective multicenter study, 107 NSCLC patients, 44 patients with V600E BRAF mutation, were treated with anti-PD-L1 immunotherapy, and no increase in ICI efficacy was observed compared to non-selected patients (44). Therefore, our study adds valuable data about WT BRAF RNA expression that might lead to different and broader patterns of enhanced proliferation signals leading to primary or secondary resistance to oncotherapy.
This study has limitations. First, it is a retrospective cross-sectional study, with a modest sample size. TMA specimens might not reflect tumor heterogeneity unambiguously, despite the relatively large size of the TMA cores (3 mm) used in triplicates. Samples were taken before the inception of targeted and immunotherapies (2006–2013), so we could not assess the predictive role of any biomarker. Due to the low number of patients with available data concerning TMB and driver mutations (KRAS, EGFR), we cannot draw conclusion in connection with these parameters and BRAF RNA expression. Prospective studies with higher case numbers are needed to comprehensively characterize the prognostic and potential predictive role of BRAF expression in LADC.
Conclusions
We found an increased expression of BRAF RNA in all stages in LADC, a critical proliferation pathway in NSCLC. We showed that a significant portion of LADCs express high levels of WT BRAF RNA. Nonetheless, high BRAF expression was associated with different outcomes. We hypothesize that the translational relevance of WT compared to the mutant BRAF expression represents a broader spectrum of clinical applications. Furthermore, our study reveals the possibility of enhancing lung cancer therapies through further evaluating the potential of targeting overexpressed BRAF pathways. Therefore, based on the low incidence of BRAF mutations reported, we propose a further evaluation of RNA expression as reflex testing for clinical assessment in LADC.
Acknowledgments
Funding: The work was supported by the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement No. 739593 and by a Momentum Research Grant from the Hungarian Academy of Sciences (No. LP-2021-14 to ZVV). ZL and ZVV acknowledge funding from the Hungarian National Research, Development and Innovation Office (OTKA #124652, OTKA #129664, and OTKA #128666, and OTKA #134751). IV was supported by the New National Excellence Program of the Ministry for Innovation and Technology from the source of the National Research, Development and Innovation Fund (No. ÚNKP-21-3-II-SE-14) and EFOP-3.6.3-VEKOP-16-2017-00009 (Project No. RRF-2.3.1-21-2022-00003, implemented with the support provided by the European Union).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-449/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-449/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-449/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-449/coif). ZVV reports that this work was supported by the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement No. 739593 and by a Momentum Research Grant from the Hungarian Academy of Sciences. ZL and ZVV acknowledge funding from the Hungarian National Research, Development and Innovation Office (OTKA #124652, OTKA #129664, and OTKA #128666, and OTKA #134751). IV was supported by the New National Excellence Program of the Ministry for Innovation and Technology from the source of the National Research, Development and Innovation Fund (No. ÚNKP-21-3-II-SE-14) and EFOP-3.6.3-VEKOP-16-2017-00009 (Project No. RRF-2.3.1-21-2022-00003, implemented with the support provided by the European Union). The authors have no other conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- van Zandwijk N, Mathy A, Boerrigter L, et al. EGFR and KRAS mutations as criteria for treatment with tyrosine kinase inhibitors: retro- and prospective observations in non-small-cell lung cancer. Ann Oncol 2007;18:99-103. [Crossref] [PubMed]
- Massarelli E, Varella-Garcia M, Tang X, et al. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res 2007;13:2890-6. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Grosse A, Grosse C, Rechsteiner M, et al. Analysis of the frequency of oncogenic driver mutations and correlation with clinicopathological characteristics in patients with lung adenocarcinoma from Northeastern Switzerland. Diagn Pathol 2019;14:18. [Crossref] [PubMed]
- Lohinai Z, Hoda MA, Fabian K, et al. Distinct Epidemiology and Clinical Consequence of Classic Versus Rare EGFR Mutations in Lung Adenocarcinoma. J Thorac Oncol 2015;10:738-46. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013;31:3327-34. [Crossref] [PubMed]
- Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014;371:2167-77. [Crossref] [PubMed]
- Mackiewicz J, Mackiewicz A. BRAF and MEK inhibitors in the era of immunotherapy in melanoma patients. Contemp Oncol (Pozn) 2018;22:68-72. [Crossref] [PubMed]
- Planchard D, Besse B, Groen HJM, et al. Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non-small cell lung cancer: an open-label, multicentre phase 2 trial. Lancet Oncol 2016;17:984-93. [Crossref] [PubMed]
- Leonetti A, Facchinetti F, Rossi G, et al. BRAF in non-small cell lung cancer (NSCLC): Pickaxing another brick in the wall. Cancer Treat Rev 2018;66:82-94. [Crossref] [PubMed]
- Negrao MV, Raymond VM, Lanman RB, et al. Molecular Landscape of BRAF-Mutant NSCLC Reveals an Association Between Clonality and Driver Mutations and Identifies Targetable Non-V600 Driver Mutations. J Thorac Oncol 2020;15:1611-23. [Crossref] [PubMed]
- Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol 2011;12:175-80. [Crossref] [PubMed]
- Flavahan WA, Gaskell E, Bernstein BE. Epigenetic plasticity and the hallmarks of cancer. Science 2017;357:eaal2380. [Crossref] [PubMed]
- Hata AN, Niederst MJ, Archibald HL, et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med 2016;22:262-9. [Crossref] [PubMed]
- Dora D, Rivard C, Yu H, et al. Neuroendocrine subtypes of small cell lung cancer differ in terms of immune microenvironment and checkpoint molecule distribution. Mol Oncol 2020;14:1947-65. [Crossref] [PubMed]
- Dora D, Rivard C, Yu H, et al. Characterization of Tumor-Associated Macrophages and the Immune Microenvironment in Limited-Stage Neuroendocrine-High and -Low Small Cell Lung Cancer. Biology (Basel) 2021;10:502. [Crossref] [PubMed]
- Bankhead P, Loughrey MB, Fernández JA, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep 2017;7:16878. [Crossref] [PubMed]
- Uhlén M, Fagerberg L, Hallström BM, et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347:1260419. [Crossref] [PubMed]
- Cardarella S, Ogino A, Nishino M, et al. Clinical, pathologic, and biologic features associated with BRAF mutations in non-small cell lung cancer. Clin Cancer Res 2013;19:4532-40. [Crossref] [PubMed]
- Chen D, Zhang LQ, Huang JF, et al. BRAF mutations in patients with non-small cell lung cancer: a systematic review and meta-analysis. PLoS One 2014;9:e101354. [Crossref] [PubMed]
- Nieto P, Ambrogio C, Esteban-Burgos L, et al. A Braf kinase-inactive mutant induces lung adenocarcinoma. Nature 2017;548:239-43. [Crossref] [PubMed]
- Planchard D, Kim TM, Mazieres J, et al. Dabrafenib in patients with BRAF(V600E)-positive advanced non-small-cell lung cancer: a single-arm, multicentre, open-label, phase 2 trial. Lancet Oncol 2016;17:642-50. [Crossref] [PubMed]
- Aramini B, Banchelli F, Bettelli S, et al. Overall survival in patients with lung adenocarcinoma harboring "niche" mutations: an observational study. Oncotarget 2020;11:550-9. [Crossref] [PubMed]
- Wiesweg M, Preuß C, Roeper J, et al. BRAF mutations and BRAF mutation functional class have no negative impact on the clinical outcome of advanced NSCLC and associate with susceptibility to immunotherapy. Eur J Cancer 2021;149:211-21. [Crossref] [PubMed]
- Tanami H, Imoto I, Hirasawa A, et al. Involvement of overexpressed wild-type BRAF in the growth of malignant melanoma cell lines. Oncogene 2004;23:8796-804. [Crossref] [PubMed]
- Smalley KS, Contractor R, Nguyen TK, et al. Identification of a novel subgroup of melanomas with KIT/cyclin-dependent kinase-4 overexpression. Cancer Res 2008;68:5743-52. [Crossref] [PubMed]
- Couraud S, Barlesi F, Fontaine-Deraluelle C, et al. Clinical outcomes of non-small-cell lung cancer patients with BRAF mutations: results from the French Cooperative Thoracic Intergroup biomarkers France study. Eur J Cancer 2019;116:86-97. [Crossref] [PubMed]
- Reiniger L, Téglási V, Pipek O, et al. Tumor necrosis correlates with PD-L1 and PD-1 expression in lung adenocarcinoma. Acta Oncol 2019;58:1087-94. [Crossref] [PubMed]
- Rajakulendran T, Sahmi M, Lefrançois M, et al. A dimerization-dependent mechanism drives RAF catalytic activation. Nature 2009;461:542-5. [Crossref] [PubMed]
- Eroglu Z, Ribas A. Combination therapy with BRAF and MEK inhibitors for melanoma: latest evidence and place in therapy. Ther Adv Med Oncol 2016;8:48-56. [Crossref] [PubMed]
- Queirolo P, Picasso V, Spagnolo F. Combined BRAF and MEK inhibition for the treatment of BRAF-mutated metastatic melanoma. Cancer Treat Rev 2015;41:519-26. [Crossref] [PubMed]
- Rudin CM, Hong K, Streit M. Molecular characterization of acquired resistance to the BRAF inhibitor dabrafenib in a patient with BRAF-mutant non-small-cell lung cancer. J Thorac Oncol 2013;8:e41-2. [Crossref] [PubMed]
- Amaral T, Sinnberg T, Meier F, et al. MAPK pathway in melanoma part II-secondary and adaptive resistance mechanisms to BRAF inhibition. Eur J Cancer 2017;73:93-101. [Crossref] [PubMed]
- Amaral T, Sinnberg T, Meier F, et al. The mitogen-activated protein kinase pathway in melanoma part I - Activation and primary resistance mechanisms to BRAF inhibition. Eur J Cancer 2017;73:85-92. [Crossref] [PubMed]
- Acosta AM, Kadkol SS. Mitogen-Activated Protein Kinase Signaling Pathway in Cutaneous Melanoma: An Updated Review. Arch Pathol Lab Med 2016;140:1290-6. [Crossref] [PubMed]
- Marchetti A, Felicioni L, Malatesta S, et al. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol 2011;29:3574-9. [Crossref] [PubMed]
- Paik PK, Arcila ME, Fara M, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol 2011;29:2046-51. [Crossref] [PubMed]
- Ding X, Zhang Z, Jiang T, et al. Clinicopathologic characteristics and outcomes of Chinese patients with non-small-cell lung cancer and BRAF mutation. Cancer Med 2017;6:555-62. [Crossref] [PubMed]
- Cui G, Liu D, Li W, et al. A meta-analysis of the association between BRAF mutation and nonsmall cell lung cancer. Medicine (Baltimore) 2017;96:e6552. [Crossref] [PubMed]
- Simanshu DK, Nissley DV, McCormick F. RAS Proteins and Their Regulators in Human Disease. Cell 2017;170:17-33. [Crossref] [PubMed]
- Dudnik E, Peled N, Nechushtan H, et al. BRAF Mutant Lung Cancer: Programmed Death Ligand 1 Expression, Tumor Mutational Burden, Microsatellite Instability Status, and Response to Immune Check-Point Inhibitors. J Thorac Oncol 2018;13:1128-37. [Crossref] [PubMed]
- Katano T, Oda T, Sekine A, et al. Five cases of BRAF V600E-mutant lung adenocarcinoma with high expression of programmed death ligand 1. Respir Med Case Rep 2020;30:101071. [Crossref] [PubMed]
- Guisier F, Dubos-Arvis C, Viñas F, et al. Efficacy and Safety of Anti-PD-1 Immunotherapy in Patients With Advanced NSCLC With BRAF, HER2, or MET Mutations or RET Translocation: GFPC 01-2018. J Thorac Oncol 2020;15:628-36. [Crossref] [PubMed]


