Customized chemotherapy in metastatic non-small cell lung cancer (NSCLC)
Introduction
Growing evidence points to the need for molecular characterization of non-small cell lung cancer (NSCLC), especially in adenocarcinomas and never smokers, for adequate identification of driver mutations or translocations that can be properly treated with targeted therapy. However, there is still a large proportion of NSCLCs for which genetic information to inform therapeutic intervention is still lacking. The benefit of chemotherapy is rather limited and almost all advanced NSCLC cases have poor prognosis with median survival of less than one year. Previous studies comparing chemotherapy with best supported care showed median survivals of between 8 and 4 months, respectively (1). Different studies of chemotherapy with cetuximab or pemetrexed as maintenance therapy have not significantly improved overall survival (2-4). In this review we will describe the significant components in DNA repair pathways that warrant investigation, with the aim of identifying a predictive model for optimal customization of chemotherapy which could translate to a meaningful improvement in survival. A BRCA1 and RAP80 Expression Customized (BREC) phase III trial (NCT00617656/GECP-BREC) and a parallel phase II study in China (BREC China, ChiCTR-TRC-12001860) are currently being performed based on the biological information available in 2007. Since then, great progress has been made in further defining DNA repair mechanisms. In this review we will summarize this important progress that has occurred whilst awaiting the results of the above mentioned trials. Figure 1 shows the design of the randomized BREC trial.
RAP80 and BRCA1 mRNA levels in customizing chemotherapy in the BREC
The BREC studies were constructed based on a Spanish Lung Cancer Group (SLGC) phase II customized trial (NCT00883480) and information that was discovered in 2007 regarding the BRCA1-A complex (BRCA1, RAP80, ABRAXAS). As commented, information which has since been reported, during the accrual of the BREC, provides the rationale for exploring the mRNA levels of other genes in the BREC patients - above all, RNF8 could play a decisive role, since, when BRCA1 and RAP80 are low, if RNF8 is still expressed this will neutralize the predictive model. Other interesting genes and associations are explained below.
Double-strand breaks (DSBs) induced by chemotherapy lead to DNA damage response (DDR): ATM-related or tyrosine kinase-driven
DNA DSBs caused by chemotherapy are repaired by two major systems: non-homologous end joining (NHEJ) and homologous recombination (HR). Upon DNA DSB introduction, the following processes occur: the histone H2AX is phosphorylated by ataxia telangiectasia mutated (ATM); the mediator of DNA damage checkpoint 1 (MDC1) binds to the phosphorylated H2AX (H2AX); ATM phosphorylates MDC1 at the region surrounding the DSB. The E3 ubiquitin ligase RING finger protein 8 (RNF8) binds to phosphorylated MDC1 at DSB sites and promotes recruitment of another E3 ubiquitin ligase RNF168; RNF8 and RNF168 conjugate Lys 63-linked ubiquitin chains onto histone H2A with their cognate E2 ubiquitin-conjugating enzyme UBC13 and induce chromatin remodeling. UBC13-RNF8/RNF168-dependent ubiquitination promotes recruitment of BRCA1 and p53-binding protein 1 (53BP1) to DSBs (5) (Figure 2). Importantly, a large proportion of BRCA1 that localizes to DSB sites is a component of the BRCA1-A complex, consisting of a BRCA1/BARD1 heterodimer, ubiquitin interacting motif (UIM)-containing protein RAP80, and adaptor protein ABRAXAS (6-9).
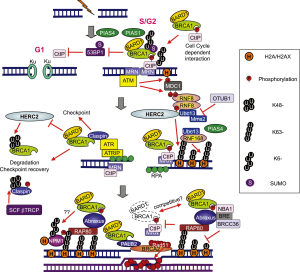
Based on this information, we performed an exploratory analysis of RAP80 and ABRAXAS mRNA levels in our previous customized phase II trial. Although the information provided by ABRAXAS was similar to that provided by RAP80, RAP80 was more significant (10). Mechanistically, loss of RAP80 suppresses recruitment of the BRCA1 complex to DNA damage sites and abrogates the DNA damage repair process at DSBs (11). It has since been discovered that the BRCA1-A complex also includes the deubiquitinating enzyme BRCC36, as well as BRCC45/BRE and MERIT40/NBA1 (5). Other groups have also demonstrated that, BRCA1 forms biochemically distinct complexes with certain other DNA damage response proteins [BRCA1-B and BRCA1-C complexes; Figure 3 (6)] in response to DSBs. The simultaneous presence of multiple distinct BRCA1 complexes at DSBs suggests a crosstalk between complexes and increases the level of complexity; for example, the BRCA1/RAP80 complex can mitigate excessive resection by CtIP (12). Although a large proportion of BRCA1 fails to be retained at DSBs upon loss of RAP80, it is possible that relocation of a small amount of BRCA1 to the DSBs via the association with other protein complexes could occur.
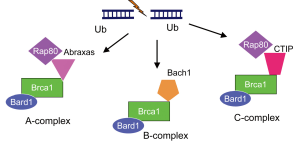
In addition, BRCA1 can be recruited to DSBs through direct binding to phosphorylated CtIP, forming the BRCA1-C complex (6) [Figure 3 (6)]. Importantly, CtIP is capable of generating limited DSB end resection without BRCA1 to promote altered NHEJ, an error-prone repair in G1 phase of the cell cycle [Figure 2 (5)]. Interestingly, the DSB end resection promoted by CtIP is inhibited by 53BP1, and BRCA1 overwhelms 53BP1 to execute the resection (13,14). In addition, 53BP1 blocks HR and sustains the growth arrest induced by BRCA1 depletion. One major function of BRCA1 and BRCA1-C complex is the suppression of 53BP1 and prolongation of the CtIP activity for DSB end resection to generate ssDNA length long enough for HR [Figure 2 (5)].
RAP80 interacts with Lys63-linked chain that is generated by UBC13-RNF8/RNF168 and brings BRCA1 to DSB sites. The overexpression of the deubiquitinating enzyme OUT domain, ubiquitin aldehyde binding 1 (OTUB1) suppresses DNA damage-dependent chromatin ubiquitination through inhibition of UBC13 activity, thus suppressing HR (15) [Figure 2 (5)].
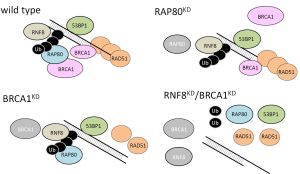
One of the major difficulties in the BREC study is that tumor cells have multiple DNA repair systems other than HR. These systems work redundantly, each operating to repair DNA in the event that other repair systems are ineffective. Recently, it has been demonstrated that inhibition of RNF8 or RNF168 activity can suppress BRCA1 independent of HR in tumor cells with low 53BP1. RNF8 is required for resistance to both irradiation and cytotoxic drugs (16). RNF8 can promote RAD51 assembly at DSB sites in BRCA1/53BP1-depleted cells (17). The model shows that in normal cells, an ubiquitin chain of RAP80, BRCA1, 53BP1 and RAD51 assembles at DSB sites. In BRCA1-depleted cells, RAP80 and 53BP1, but not RAD51, assemble at DSB sites. In RAP80-depleted cells, a small subset of BRCA1 protein, 53BP1 and RAD51 assemble at DSB sites. However, in RNF8/BRCA1-depleted cells or in RNF8/BRCA1/53BP1-depleted cells, RAD51 and RAP80 do not assemble at DSB sites (17) (Figure 4).
RNF8 displays dual non-catalytic and catalytic activities responsible for chromatin decondensation and histone ubiquitylation, respectively. An RNF8 dimer is recruited to a DSB by binding to phosphorylated MDC1. The recruited RNF8 dimer binds to the chromodomain helicase DNA-binding protein 4 (CHD4) in a phospho-independent manner, resulting in local chromatin decondensation, which permits enhanced ubiquitin conjugation at DSBs and association of RNF168 and BRCA1 (18). In addition, the ubiquitin-selective valosin-containing protein (VCP) is recruited by RNF8 and plays a critical role in mediating the recruitment of downstream repair factors. VCP stimulates 53BP1 recruitment (18).
The function of RNF8 could be vital to chemoresistance. The HECT type E3 ligase (HERC2), a large 4834-amino acid protein, interacts with the FHA domain of RNF8 in a phosphorylation-dependent manner, facilitating assembly of the RNF8/UBC13 complex (19) [Figure 2 (5)]. Therefore, analysis of HERC2 and RNF8 could be of potential relevance in interpreting the results of the BREC. Interestingly, HERC2 can degrade BRCA1 (19). Also, nucleophosmin (NPM1) is recruited to DSBs in a manner dependent on the RNF8/RNF168-mediated ubiquitin conjugates (20).
PIAS1 and PIAS4 are recruited to DSBs. Depletion of PIAS1 or PIAS4 reduces the proportion of cells displaying BRCA1 accumulation and increase BRCA1 staining intensity at DSBs, increasing sensitivity to irradiation or cisplatin (21,22). Recruitment of RNF168 is impaired only in PIAS4- but not in PIAS1-depleted cells. 53BP1 recruitment does not require BRCA1 or PIAS1 but does require PIAS4 (21,22) [Figure 2 (5)]. This important finding indicates the importance of examining BRCA1 levels together with those of PIAS1, as well as 53BP1 together with PIAS4 levels. High levels of PIAS4, PP2A/C and BRCA1mRNA were all independent markers of shorter PFS in EGFR-mutant non-small-cell lung cancer (NSCLC) patients treated with erlotinib (23). Along the same lines, low levels of BRCA1, PIAS1 and PIAS4 were independent markers of poor survival in gastric cancer patients receiving docetaxel as second-line treatment (24). BRCA1 was found to be a differential modulator of chemosensitivity, inducing a 10-1000-fold increase in resistance to several DNA-damaging agents, especially those that give rise to DSBs. In contrast, BRCA1 induced a more than 1000-fold increase in sensitivity to paclitaxel, docetaxel and vinorelbine (25,26).
RNF8 could establish a bridge between HR and the NHEJ repair. RNF8 regulates the abundance of the NHEJ repair protein KU80 at sites of DNA damage. RNF8 depletion results in prolonged retention of KU80 at damage sites and impaired NHEJ (27) [Figure 2 (5)]. Therefore, we can assume that RNF8 depletion is important not only in enhancing the cytotoxic effect of chemotherapy, as described above, but also in impairing repair by NHEJ. On the other hand, NHEJ can function well in the presence of normal RNF8, which may contribute to the failure to predict outcome in the customized model of the BREC. Therefore, analysis of the BREC study can be re-interpreted according to expression of RNF8. In tumors with adequate RNF8 function, expression of Ligase IV could be a major determinant of shorter PFS. DNA Ligase IV is responsible for sealing of DSBs during NHEJ, which is one of the primary mechanisms of DSB repair and is active throughout the cell cycle. During NHEJ, KU70/KU80 heterodimer binds to the DNA ends and recruits proteins, such as DNA-PKcs, Artemis, and Pol, to the repair site, resulting in end-processing followed by Ligase IV, XRCC4 and XLF complex-mediated ligation (28). NHEJ plays a major role in resistance to chemotherapy and radiotherapy. DNA PKcs have been associated with radioresistance in lung cancer cell lines (29). Metnase is a recently described fusion protein composed of a SET histone methylase domain and Transposase nuclease domain. Metnase enhances NHEJ. Both the SET histone methylase domain and the Transposase nuclease domain are essential for the enhancement of DSB repair (30). Metnase is overexpressed in acute leukemia (31), causing resistance to chemotherapy. Decreasing metnase enhanced cisplatin sensitivity in a lung cancer A549 xenograft (32).
PPP2R2A is also a critical effector of HR through modulation of ATM phosphorylation. PPP2R2A-depleted cells dramatically increase sensitivity to PARP inhibitors. Interestingly, PPP2R2A mRNA is commonly downregulated in NSCLC (33). We have previously observed that in EGFR-mutant NSCLC patients treated with erlotinib, high levels of PP2A/C mRNA significantly increased the hazard ratio for PFS in a multivariate model (23).
Modulator of apoptosis protein 1 (MOAP-1) is a Bax-interacting protein whose knockdown inhibits apoptosis. MOAP-1 association with Bax promotes Bax mitochondrial translocation and activation. The BH3-only proteins, like BIM or BID, serve as sentinels for the initiation of apoptosis by modulating the functions of multi-domain pro-survival (Bcl-2, Mcl-1, and others) or pro-apoptotic members like Bax, involved in regulating the mitochondrial outer membrane permeability (MOMP). MOAP-1 is highly enriched in mitochondria and is considered to act as an effector to facilitate apoptotic signaling of Bax in mitochondria. The intrinsic or mitochondrial programmed cell death pathway leads to the activation of caspase-9 and then caspase-3 (34). MOAP-1 degradation is inhibited by Trim39, a member of the tripartite motif (TRIM) family. Trim39 overexpression enhances etoposide-induced, Bax-mediated apoptosis through stabilization of MOAP-1 (35). Trim39 mRNA is highly expressed in the testis. The Trim 39 gene is located in the MHC class I region of genes within chromosome 6p21-23 (36). There is a mechanistic reason for this finding, since caspase-3 cleaves MDC1, separating the BRCT and FHA domains of MDC1, thus abrogating DNA damage repair (37). These observations prompt us to speculate that BIM mRNA levels could be crucial in inducing apoptosis and that downstream effectors, such as MOAP-1, Trim39 and caspase-3, could play an important role.
DNA damage checkpoint (DDC) signaling on DNA replication
In addition to homologous recombination and NHEJ, the genotoxic stress induced by chemotherapy also causes replication stress (38). This DDC pathway is less well known. The DNA repair scaffolding proteins Slx4 and Rtt107 prevent aberrant activation of DDC signaling by lesions generated during DNA replication. On replication stress, Saccharomyces cerevisiae cells lacking Slx4 and Rtt107 show hyperactivation of the downstream DDC kinase Rad53. The Slx4 or Rtt107 complex counteracts the checkpoint adaptor Rad9 by physically interacting with Dpb11 and phosphorylated histone H2A (39). It is hypothesized that modulation of Rad53 activation occurs by a DAMP (dampens checkpoint adaptor-mediated phosphor-signaling) (39).
It has recently been described that RNF126 is highly expressed in a subset of breast cancer cell lines and negatively correlates with p21 expression levels. RNF126 targets p21 for ubiquitin-mediated degradation (40).
DNA damage response (DDR) independent of ATM
Phosphoproteomic analysis have found that several kinases can be involved in DDR, with extensive crosstalk between them. One of the most important could be c-ABL. c-ABL is a non-receptor tyrosine kinase that is upregulated following irradiation, cisplatin and other drugs. c-ABL interacts with ATM and DNA-PK. c-ABL activated by irradiation mediates phosphorylation of PI3K and mTOR, leading to the inhibition of kinase activity (41).
c-ABL is a transducer in the process of apoptosis in response to DNA damage. It is a member of the Src family of non-receptor tyrosine kinases. Under normal conditions, c-ABL is inactive and sequestered into the cytoplasm by binding to the 14-3-3 protein. Upon DNA damage, c-Jun N-terminal kinase (JNK) is activated, phosphorylating 14-3-3 at the binding site to c-ABL, which releases c-ABL, which is localized in the nucleus and is activated by phosphorylation by ATM. Of great interest is that YAP1 is a direct substrate of c-ABL, and DNA damage stabilizes YAP1 in a c-ABL kinase-dependent manner. Then, the phosphorylated YAP1 binds to p73 and is selectively recruited onto the Bax promoter to induce apoptosis (42). The Hippo signaling pathway is a novel tumor suppressor pathway, and the downstream effect of the Hippo signaling cascade is to phosphorylate and inactivate YAP1 and its paralog TAZ. YAP1 and TAZ overexpression has been observed in NSCLC, conferring poor prognosis (43,44). It is interesting that YAP1 can induce apoptosis (Bax) via c-ABL.
Intriguingly, reinforcing the role of c-ABL, it has recently been reported that overexpression of AXL causes resistance to cisplatin by inhibiting c-ABL/p73 signal (45). This allows us to reason that, since AXL is an effector of the YAP-TAZ pathway (when HIPPO is off) and can induce abrogation of c-ABL, disrupting the association with p73β (45). However, the previous work has demonstrated that c-ABL enhances apoptosis via activating YAP1 (42). This apparent contradiction can only be explained by requiring the Wnt pathway to be active since beta-catenin is then linked to YAP1 and may hamper YAP’s transcriptional program, including activation of AXL. On these grounds, also recently, YAP1 and TAZ have been observed to be coupled with beta-catenin, and the degradation of YAP1 and TAZ is avoided when the Wnt pathway is active, which abrogates the beta-catenin destruction complex (AXIN1, GSK3, APC) (46,47). Binding of the Wnts to their receptors inactivates this complex, leading to accumulation and nuclear translocation of beta-catenin (48). Also, paradoxically, in melanomas with BRAFV600E, the efficacy of the BRAF inhibitor PLX4720 is increased when beta-catenin is present, and this is achieved by eliminating AXIN1 (49).
Beta-catenin-independent signaling pathways
In addition to the FZD receptors, the Wnt receptors ROR1 and ROR2 also contribute to cancer proliferation (48). Wnt5A is the ligand for ROR1 (50). ROR1 repression inhibits lung adenocarcinoma regardless of EGFR status. ROR1 abrogates ASK1, which can lead to abrogation of BIM (51). In the EURTAC study, higher levels of ROR1 mRNA correlated significantly with poor survival.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Rapp E, Pater JL, Willan A, et al. Chemotherapy can prolong survival in patients with advanced non-small-cell lung cancer--report of a Canadian multicenter randomized trial. J Clin Oncol 1988;6:633-41. [PubMed]
- Pirker R, Pereira JR, Szczesna A, et al. Cetuximab plus chemotherapy in patients with advanced non-small-cell lung cancer (FLEX): an open-label randomised phase III trial. Lancet 2009;373:1525-31. [PubMed]
- Ciuleanu T, Brodowicz T, Zielinski C, et al. Maintenance pemetrexed plus best supportive care versus placebo plus best supportive care for non-small-cell lung cancer: a randomised, double-blind, phase 3 study. Lancet 2009;374:1432-40. [PubMed]
- Paz-Ares L, de Marinis F, Dediu M, et al. Maintenance therapy with pemetrexed plus best supportive care versus placebo plus best supportive care after induction therapy with pemetrexed plus cisplatin for advanced non-squamous non-small-cell lung cancer (PARAMOUNT): a double-blind, phase 3, randomised controlled trial. Lancet Oncol 2012;13:247-55. [PubMed]
- Ohta T, Sato K, Wu W. The BRCA1 ubiquitin ligase and homologous recombination repair. FEBS Lett 2011;585:2836-44. [PubMed]
- Wang B, Matsuoka S, Ballif BA, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 2007;316:1194-8. [PubMed]
- Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 2007;316:1202-5. [PubMed]
- Sobhian B, Shao G, Lilli DR, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 2007;316:1198-202. [PubMed]
- Yan J, Kim YS, Yang XP, et al. The ubiquitin-interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res 2007;67:6647-56. [PubMed]
- Rosell R, Perez-Roca L, Sanchez JJ, et al. Customized treatment in non-small-cell lung cancer based on EGFR mutations and BRCA1 mRNA expression. PLoS One 2009;4:e5133. [PubMed]
- Wu J, Liu C, Chen J, et al. RAP80 protein is important for genomic stability and is required for stabilizing BRCA1-A complex at DNA damage sites in vivo. J Biol Chem 2012;287:22919-26. [PubMed]
- Coleman KA, Greenberg RA. The BRCA1-RAP80 complex regulates DNA repair mechanism utilization by restricting end resection. J Biol Chem 2011;286:13669-80. [PubMed]
- Bunting SF, Callén E, Wong N, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 2010;141:243-54. [PubMed]
- Bouwman P, Aly A, Escandell JM, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol 2010;17:688-95. [PubMed]
- Nakada S, Tai I, Panier S, et al. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 2010;466:941-6. [PubMed]
- Sy SM, Jiang J, Dong SS, et al. Critical roles of ring finger protein RNF8 in replication stress responses. J Biol Chem 2011;286:22355-61. [PubMed]
- Nakada S, Yonamine RM, Matsuo K. RNF8 regulates assembly of RAD51 at DNA double-strand breaks in the absence of BRCA1 and 53BP1. Cancer Res 2012;72:4974-83. [PubMed]
- Luijsterburg MS, Acs K, Ackermann L, et al. A new non-catalytic role for ubiquitin ligase RNF8 in unfolding higher-order chromatin structure. EMBO J 2012;31:2511-27. [PubMed]
- Wu W, Sato K, Koike A, et al. HERC2 is an E3 ligase that targets BRCA1 for degradation. Cancer Res 2010;70:6384-92. [PubMed]
- Koike A, Nishikawa H, Wu W, et al. Recruitment of phosphorylated NPM1 to sites of DNA damage through RNF8-dependent ubiquitin conjugates. Cancer Res 2010;70:6746-56. [PubMed]
- Morris JR, Boutell C, Keppler M, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 2009;462:886-90. [PubMed]
- Galanty Y, Belotserkovskaya R, Coates J, et al. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 2009;462:935-9. [PubMed]
- Rosell R, Molina MA, Costa C, et al. Pretreatment EGFR T790M mutation and BRCA1 mRNA expression in erlotinib-treated advanced non-small-cell lung cancer patients with EGFR mutations. Clin Cancer Res 2011;17:1160-8. [PubMed]
- Wei J, Costa C, Ding Y, et al. mRNA expression of BRCA1, PIAS1, and PIAS4 and survival after second-line docetaxel in advanced gastric cancer. J Natl Cancer Inst 2011;103:1552-6. [PubMed]
- Quinn JE, James CR, Stewart GE, et al. BRCA1 mRNA expression levels predict for overall survival in ovarian cancer after chemotherapy. Clin Cancer Res 2007;13:7413-20. [PubMed]
- Quinn JE, Kennedy RD, Mullan PB, et al. BRCA1 functions as a differential modulator of chemotherapy-induced apoptosis. Cancer Res 2003;63:6221-8. [PubMed]
- Feng L, Chen J. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat Struct Mol Biol 2012;19:201-6. [PubMed]
- Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat Struct Mol Biol 2010;17:410-6. [PubMed]
- Sirzén F, Nilsson A, Zhivotovsky B, et al. DNA-dependent protein kinase content and activity in lung carcinoma cell lines: correlation with intrinsic radiosensitivity. Eur J Cancer 1999;35:111-6. [PubMed]
- Beck BD, Lee SS, Williamson E, et al. Biochemical characterization of metnase’s endonuclease activity and its role in NHEJ repair. Biochemistry 2011;50:4360-70. [PubMed]
- Wray J, Williamson EA, Sheema S, et al. Metnase mediates chromosome decatenation in acute leukemia cells. Blood 2009;114:1852-8. [PubMed]
- Williamson EA, Damiani L, Leitao A, et al. Targeting the transposase domain of the DNA repair component Metnase to enhance chemotherapy. Cancer Res 2012;72:6200-8. [PubMed]
- Kalev P, Simicek M, Vazquez I, et al. Loss of PPP2R2A inhibits homologous recombination DNA repair and predicts tumor sensitivity to PARP inhibition. Cancer Res 2012;72:6414-24. [PubMed]
- Del Gaizo Moore V, Brown JR, Certo M, et al. Chronic lymphocytic leukemia requires BCL2 to sequester prodeath BIM, explaining sensitivity to BCL2 antagonist ABT-737. J Clin Invest 2007;117:112-21. [PubMed]
- Huang NJ, Zhang L, Tang W, et al. The Trim39 ubiquitin ligase inhibits APC/CCdh1-mediated degradation of the Bax activator MOAP-1. J Cell Biol 2012;197:361-7. [PubMed]
- Lee SS, Fu NY, Sukumaran SK, et al. TRIM39 is a MOAP-1-binding protein that stabilizes MOAP-1 through inhibition of its poly-ubiquitination process. Exp Cell Res 2009;315:1313-25. [PubMed]
- Solier S, Pommier Y. MDC1 cleavage by caspase-3: a novel mechanism for inactivating the DNA damage response during apoptosis. Cancer Res 2011;71:906-13. [PubMed]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell 2007;28:739-45. [PubMed]
- Ohouo PY, Bastos de Oliveira FM, Liu Y, et al. DNA-repair scaffolds dampen checkpoint signalling by counteracting the adaptor Rad9. Nature 2013;493:120-4. [PubMed]
- Zhi X, Zhao D, Wang Z, et al. E3 ubiquitin ligase RNF126 promotes cancer cell proliferation by targeting the tumor suppressor p21 for ubiquitin-mediated degradation. Cancer Res 2013;73:385-94. [PubMed]
- Bensimon A, Aebersold R, Shiloh Y. Beyond ATM: the protein kinase landscape of the DNA damage response. FEBS Lett 2011;585:1625-39. [PubMed]
- Levy D, Adamovich Y, Reuven N, et al. Yap1 phosphorylation by c-Abl is a critical step in selective activation of proapoptotic genes in response to DNA damage. Mol Cell 2008;29:350-61. [PubMed]
- Wang Y, Dong Q, Zhang Q, et al. Overexpression of yes-associated protein contributes to progression and poor prognosis of non-small-cell lung cancer. Cancer Sci 2010;101:1279-85. [PubMed]
- Zhou Z, Hao Y, Liu N, et al. TAZ is a novel oncogene in non-small cell lung cancer. Oncogene 2011;30:2181-6. [PubMed]
- Hong J, Peng D, Chen Z, et al. ABL regulation by AXL promotes cisplatin resistance in esophageal cancer. Cancer Res 2013;73:331-40. [PubMed]
- Azzolin L, Zanconato F, Bresolin S, et al. Role of TAZ as mediator of Wnt signaling. Cell 2012;151:1443-56. [PubMed]
- Rosenbluh J, Nijhawan D, Cox AG, et al. β-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell 2012;151:1457-73. [PubMed]
- Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer 2013;13:11-26. [PubMed]
- Biechele TL, Kulikauskas RM, Toroni RA, et al. Wnt/β-catenin signaling and AXIN1 regulate apoptosis triggered by inhibition of the mutant kinase BRAFV600E in human melanoma. Sci Signal 2012;5:ra3. [PubMed]
- Zhang S, Chen L, Cui B, et al. ROR1 is expressed in human breast cancer and associated with enhanced tumor-cell growth. PLoS One 2012;7:e31127. [PubMed]
- Yamaguchi T, Yanagisawa K, Sugiyama R, et al. NKX2-1/TITF1/TTF-1-Induced ROR1 is required to sustain EGFR survival signaling in lung adenocarcinoma. Cancer Cell 2012;21:348-61. [PubMed]


