Genetic alteration profile of EGFR-mutant resected IIB–IIIA stage NSCLC and correlation to clinical outcomes
Introduction
Lung cancer accounts for about one-fifth of all cancer mortality and remains the leading cause of cancer-related death (1). The major histological subset of lung cancer is non-small cell lung cancer (NSCLC), which constitutes 85% of all lung cancer cases (2). Radical resection is the gold standard for the treatment for early-stage NSCLC. But only 20–25% of newly diagnosed NSCLC patients are suitable for surgery (3). After radical resection, stage IA NSCLC patient can achieve a good clinical outcome with a 5-year survival rate of 65% (4). While for NSCLC staged at IIB–IIIA, recurrence often takes place in various sites after primary tumor removal, and the prognosis is unfavorable with a 5-year survival rate ranging from 19% to 25% (5,6). The reason for this frequent recurrence and poor prognosis of stage IIB–IIIA NSCLC is undefined.
Some studies explored the relationship of driver gene alterations, especially that of epidermal growth factor receptor (EGFR) mutations and anaplastic lymphoma kinase (ALK) fusion with the clinical outcome of early-stage NSCLC. One study demonstrated that EGFR mutations, as well as ALK fusion, were favorable prognostic factors of time to recurrence in resectable NSCLC (7); however, another study found that ALK fusion had a different impact on resectable NSCLC of different stages. In stage IA, ALK fusion-positive NSCLC, patients had longer disease-free survival (DFS) period, while shorter DFS was present in stage IIIA or locally advanced NSCLC harboring ALK fusion (8). On the contrary, EGFR mutation was proven to be a negative prognostic indicator of DFS in postoperative NSCLC in another study (9). While a meta-analysis concluded that EGFR mutant status had no significant impact on the DFS of patients with radical resected NSCLC (4). However, EGFR exon 19 deletions might be a negative predictor for prognosis (4). Based on these conflicting results, no definite conclusion about which genomic changes are associated with the prognosis of resectable NSCLC has been reached. Further research is needed to illustrate the association between genetic alterations and the DFS of early-stage NSCLC.
Despite EGFR mutations and ALK fusion, many other driver genes’ alterations have been confirmed to cause poor prognosis of NSCLC (10,11). This negative impact of concurrent genetic alterations on the survival period was often correlated to tyrosine kinase inhibitors (TKIs) resistance in EGFR sensitive NSCLC. Since EGFR-TKI is applied in advanced NSCLC, the study of the effect of concurrent driver gene mutation on clinical outcome majorly focused on late-stage cases. To the best of our knowledge, only one study from Canada proved that multiple driver-gene alterations showed poorer DFS of postoperative NSCLC (9). A study from China just presented the mutation profile of resected EGFR-mutated lung adenocarcinoma (12). Few pieces of research have addressed the profile of concurrent mutant genes in EGFR positive resectable lung cancer and its effect on recurrence after tumor removal.
We hypothesized that concurrent mutations of EGFR with other driver genes might play a role in the recurrence and negative clinical outcome of resectable NSCLC. Therefore, we carried out the current study to illustrate the prevalence of EGFR co-mutation in resectable NSCLC and its effect on DFS as well as other clinicopathological features. Also, we analyzed the effect of each concurrent mutant gene on DFS to clarify which one is critical. These critical mutant genes may serve as prognostic biomarkers in terms of recurrence for resectable NSCLC and can be used to screen high-risk patients to implement intensive surveillance after tumor resection.
Methods
Patients and samples
From June 2016 to June 2018, consecutive patients who were diagnosed as NSCLC and underwent a thoracic operation in the First Affiliated Hospital of Guangzhou Medical University Hospital were screened retrospectively. The baseline and clinicopathological information of those who met the including criteria were collected. Besides, Formalin-Fixed and Paraffin-Embedded (FFPE) sections loading tumor tissues were gathered to perform further detection. Patients were followed up every three months. Computed tomography on chest and abdomen was conducted for them to examine tumor recurrence on each visit. Informed consents were obtained from all participants or their authorized relatives. This study was carried out in compliance with the Declaration of Helsinki Principles and was approved by the Ethics Committee of the First Affiliated Hospital of Guangzhou Medical University (No. CCTC1501).
Targeted next-generation sequencing (NGS)
The DNA extraction was performed using the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany). Targeted NGS was performed to detect somatic mutations within each sample using a 285-gene panel on the Ion Torrent platform. These 285 genes included within the panel were listed in http://cdn.amegroups.cn/static/application/9be230dec1cfedbde411e86d6d2ac4de/10.21037tlcr.2019.10.19-1.pdf.
Data processing
Somatic mutation calling was performed using VarDict. The somatic mutations in each patient’s sample were annotated with ANNOVAR. Mutations with an incidence higher than 1% in esp6500, gnomAD and ExAC database were filtered out as germline mutations. The remaining somatic mutations with ANNOVAR’ annotations were converted to a MAF file using maftools. The oncoprint was drawn using nonsynonymous mutations with maftools.
Statistical analysis
Metric data were analyzed by the Mann-Witney U test. Fisher's exact test or Chi-square test was used to compare the categorical data between two groups. Kaplan-Meier method was used to analyze the DFS. The log-rank test was applied to compare DFS curves stratified by different clinical or genetic factors. To analyze factors correlating to DFS, multivariable Cox proportional hazards model was applied. Statistical analyses were carried out using the software package SPSS 22.0 (IBM Corp., Somers, NY, USA). Statistical significance was defined as two-tailed P values<0.05. Forest plot of multivariable Cox proportional hazards was depicted using Graphpad Prism 8.
Results
Demographic and clinicopathological characteristics
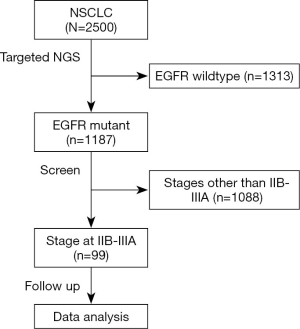
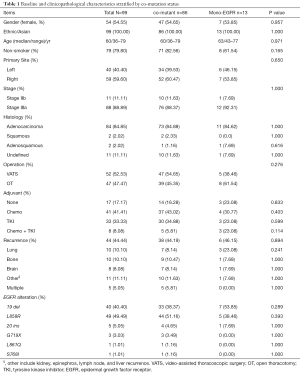
Targeted NGS was performed on those who were confirmed by histology as NSCLC (n=2,500). Among them, 1,187 tumors were defined as harboring EGFR mutations, from which 99 patients staged at IIB–IIIA were enrolled in this study. The flowchart of the patient-selection process was shown in Figure 1. Concurrent driver gene mutations were detected in 86 participants. Demographic and clinicopathological information stratified by concurrent driver gene mutation statuses were shown in Table 1. Among 99 NSCLC participants enrolled in the current study, 54.55% were females. The ages of these subjects ranged from 36 to 79 years old, and the median age was 60 years old. Most of them were diagnosed with NSCLC staged at IIIa (88.89%) with predominant histology of adenocarcinoma (84.85%). About 52.53% received video-assisted thoracoscopic surgery (VATS), while the remaining 47.47% received open thoracotomy (OT). EGFR mutation detection found that 89 tumor samples harbored EGFR common mutations, i.e., exon 19 deletion or p.L858R. Five harbored EGFR exon 20 insertions, and five were other types of rare mutations. After the operation, most patients (82.83%) received adjuvant therapy using chemotherapeutic agents and/or EGFR-TKI. Forty-four patients presented with tumor recurrence by the last follow-up. Lung and bone were the most frequent recurrent sites, followed by the brain. And tumors re-occurred in multiple sites in five patients.
Patients with concurrent driver gene alterations showed larger fractions of non-smokers and right-sided lung cancer. Besides, patients with co-occurring mutations had less frequent lung recurrence, more p.L858R mutation, and less frequent EGFR exon 19 deletions. Nevertheless, no statistical difference was found in baseline and clinicopathological features between the two groups of patients with or without concurrent driver mutations (P≥0.114) as shown in Table 1. In other words, the presence of concurrent driver gene mutations did not have a significant impact on the baseline and clinicopathological characteristics of EGFR mutant NSCLC.
Full table
Co-occurring mutations of driver genes
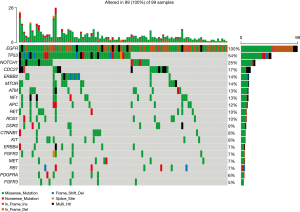
Except for EGFR, a total of 34 mutant driver genes were identified in this study. The median number of mutated driver genes for each sample was 2, ranging from 0 to 12. An overview of EGFR mutations and the top 19 most frequent concurrent driver genes’ mutations in 99 patients were depicted in Figure 2. TP53 (53.54%) was the most common mutant driver gene accompanying EGFR mutation in stage IIb, and IIIa resected NSCLC, followed by NOTCH1 mutations (25.25%). No significant difference was found in the number and frequency of concurrent driver genes between patients with different recurrence sites. Concurrent driver genes did not have a specific effect on NSCLC recurrence sites.
DFS analysis
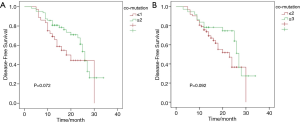
Among 99 patients, 2 were out of contact within 12 months, while 5 were out of contact 1 year after surgery. The remaining 92 patients were caught up till this research was due. The DFS period of the patients in this study ranged from 3 to 34 months, with a median DFS of 15 months. Univariate analysis was performed to explore factors that may influence DFS. These analyzed factors, as shown in Figure 3, included operation techniques, EGFR common mutations, EGFR rare mutations, adjuvant agents, and concurrent mutation status. Except for the adjuvant regimen, the univariate analysis did not find any of these factors have a statistically significant impact on DFS. Patients who received adjuvant EGFR-TKI after tumor resection had the longest DFS, followed by the combination of chemotherapy and TKI and then by chemotherapy. Patients who did not get any adjuvant therapy had the poorest DFS (P=0.013). To find out whether DFS was correlated to the number of concurrent driver genes, we further analyzed DFS based on the number of mutant driver genes for each sample. The number was stratified with breakpoints of 1 and 2 in all patients. Patients with samples harboring only one concurrent mutant driver gene had shorter DFS than those harboring more than two mutant driver genes (P=0.072) and those with less than two concurrent driver genes showed shorter DFS than those with more than three concurrent genes (P=0.092). Still, no statistical difference was found, as shown in Figure S1.
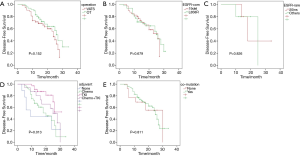
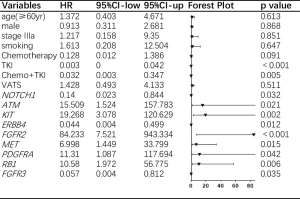
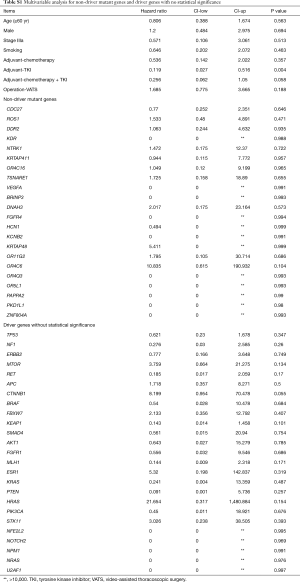
To further analyze the effect of each concurrent mutant driver gene on DFS, we carried out multivariate analysis adjusted by gender, age, stage, smoking history, operation technique, and adjuvant agents. As depicted in Figure 4, nine concurrent driver genes had a statistically significant effect on the clinical outcome of resected NSCLC with EGFR mutations. The mutant genotype of ATM, KIT, FGFR2, MET, PDGFRA, RB1, and wildtype NOTCH1, ERBB4, FGFR3 were independent risk factors for shorter DFS period. Detailed information about the HR and P value for each driver gene with a statistically significant effect on DFS was shown in Figure 3, while the HRs and P values of those driver genes with no significant effect on DFS was listed in Table S1.
Full table
In addition to concurrent driver genes, we also analyzed the effect of co-occurring non-driver gene mutations on the DFS of EGFR mutant NSCLC. In total, 22 mutant non-driver genes were detected in this study. No significant difference was found in the number and frequency of concurrent non-driver genes between patients with different recurrence sites. None of the concurrent non-driver genes was statistically significant when the effect on DFS was considered. Detailed results of multivariate analysis for non-driver gene mutations were included in Table S1.
In summary, mutations of ATM, KIT, FGFR2, MET, PDGFRA, RB1, and wildtype NOTCH1, ERBB4, FGFR3 were independent risk factors for the recurrence of resected EGFR mutant NSCLC while the number and frequency of concurrent genes were not correlated to the sites of NSCLC recurrence.
Discussion
The clinical outcome of patients with NSCLC staged at IIB-IIIA was unfavorable even after radical resection (5,6). High incidence of recurrence reduced the benefits of radical resection, especially in local advanced NSCLC. The underlying mechanism of this high recurrence rate and short DFS was unclear. Some studies have proved that co-occurring driver gene mutations were negatively correlated to clinical outcomes in advanced NSCLC (10,11). Few studies have explored the effect of concurrent driver gene mutations on the prognosis of resectable NSCLC. The current study clarified the relationship of driver gene mutations (other than EGFR) and the DFS of NSCLC harboring EGFR mutation. We also figured out several genetic events that were independent risk factors for short DFS period. This showed that the high incidence of recurrence of stage IIB-IIIA EGFR mutant NSCLC after resection was partly due to concurrent genetic alterations. The genetic variations with statistical significance may serve as prognostic indicators to guide clinical practice and help determine which patient needs more intensive surveillance after tumor resection.
EGFR mutations occur in about 50% NSCLC in Chinese. And the most frequent type of EGFR alterations in NSCLC are exon 19 deletion and p.L858R, which account for 90% of NSCLC (13,14). In the current study, the profile of EGFR mutation was comparable to that in the Chinese population from other studies. Therefore, the enrolled EGFR mutant NSCLC patients were representative when fractions of EGFR mutation subtypes were considered.
Co-occurring genetic alterations are common in patients with EGFR mutant NSCLC and play a role in oncogenesis and primary resistance to EGFR-TKI, which challenged the single-driver theory of NSCLC with EGFR sensitive mutation (15). According to our results, 86.87% of resected EGFR mutant NSCLC harbored one or more aberrant driver genes. The most frequent mutant driver gene accompanying EGFR was TP53, followed by NOTCH1. Besides, it seemed like the more the concurrent driver genes, the better the DFS of EGFR mutant NSCLC. Nevertheless, the further univariate analysis found that the presence of concurrent driver genes’ alterations or their numbers did not have a statistically significant effect on DFS. We assumed that this negative result might be owing to the heterogeneity of concurrent mutant genes among different samples. Given that different concurrent mutant genes may have contrasting effects on DFS taking them to evaluate their correlation to DFS may be misleading. Therefore, we performed a multivariable analysis to assess the effect of each mutant driver gene on DFS.
Multivariable analysis of our research discovered that mutations of ATM, KIT, FGFR2, MET, PDGFRA, RB1, and wildtype NOTCH1, ERBB4, FGFR3 were independent risk factors for the recurrence of resected EGFR mutant NSCLC. These genes involved in four signaling pathways which play important roles in the oncogenesis of NSCLC (16). FGFR2, ERBB4, FGFR3, PDGFRA, MET, and KIT are members of the RTK/RAS signaling pathway which contributes to the proliferation and cell survival of tumors. ATM is a key factor of the p53 pathway and NOTCH1, Notch pathway, while RB1 is a critical gene of the cell cycle pathway (16). NSCLC harboring EGFR mutations were accompanied by statistically significant driver genes, and substantial heterogeneity in these driver events was present between tumors.
Among the significantly mutant nine genes, NOTCH1 had the strongest association with DFS. The function of Notch signaling is extraordinarily complicated and can be opposite in different context (17). The role of Notch in NSCLC is still unclear. One study showed that activation of NOTCH1 promoted epithelial–mesenchymal transition (EMT) in lung cancer cell lines that have acquired-resistance toward gefitinib (18). Another study proved that NOTCH1 contributed to EGFR-TKI acquired resistance in NSCLC both in vitro and in vivo (19). These researches showed that normal expression of NOTCH1 might contribute to the poor clinical outcome of EGFR-mutant resected NSCLC. In line with this, from our study, it can be inferred that harmful mutation of NOTCH1 impeded NSCLC recurrence.
Multiple membrane proteins of RTK/RAS signaling pathway serve as alternative receptors for EGFR to activate RTK/RAS signals and lead to EGFR-TKI resistance. For example, FGFR2 signaling activation induced by EGFR-TKI was showed to be a new and rapid way of acquired resistance to TKI in EGFR sensitive cell lines by an in vitro study (20). The overexpression of FGFR2 in tumor samples might be an early phenomenon in the pathogenesis of NSCLC and could be a potential novel target for lung cancer (21). MET signaling activation is associated with oncogenesis and tumor progression, promoting cellular invasiveness, which leads to tumor metastasis (22). c-KIT was considered as a biomarker for cancer stem cell. High-level expression and strong staining of c-KIT was shown to be a negative prognostic factor of clinical outcome for NSCLC as well as small cell lung cancer (23,24). While in our research, mutation of KIT was an independent risk factor for shorter DFS in resected EGFR mutant NSCLC. Agents targeting the KIT signaling pathway may allow patients to harbor concurrent EGFR and KIT mutations to survive longer. RB1 was suggested by other studies to be associated with EGFR-TKI resistance and small cell lung cancer transformation of NSCLC (25,26), which may also contribute to the poor clinical outcomes of resected NSCLC.
Our results showed that these nine genes were closely correlated to the DFS of resected NSCLC. It is reasonable to include these genes into the NSCLC genetic diagnostic panel to predict clinical outcomes. However, the expression level of these genes was not defined, so we could not determine based on this research the actual contribution of each gene to DFS.
Adjuvant treatment with EGFR-TKIs demonstrates superior efficacy, compared with conventional chemotherapy, for patients with EGFR mutant resectable NSCLC (27). Besides, patients in the locally advanced stage had benefited more than those in the early-stage cases. Consistently, the current study discovered a significant difference in DFS between patients receiving different adjuvant agents. Patients receiving adjuvant EGFR-TKIs had longer DFS than those who obtained chemotherapy. Nevertheless, the DFS of patients who received a combination of chemotherapy and TKI was shorter than those who receive TKI solely. This may be a consequence of the small number size and more severe disease of the combination group, which caused a bias against patients using joint regimen.
The current study explored the profile of co-occurring mutations in resected NSCLC harboring EGFR mutations, and for the first time, we defined several genetic events associated with DFS. These events can be included in NSCLC mutation detection panel and used as biomarkers to screen recurrence high-risk patients in clinical practice. The obvious shortcoming of this study was the small sample size. Consequently, the profile of concurrent altered genes was not comprehensive, and some significant aberrated genes with low incidence failed to be included, like ALK and ROS1 fusion. Besides, the expression level of these significantly mutant genes remained unclarified. As a result, we do not know the actual effect of these pathological mutations on the expression of these genes. Large cohort studies are needed to supply a comprehensive scenario of EGFR concurrent genetic alterations in resected NSCLC and to define the impact of these alterations on clinical outcomes as well as the underlying mechanisms.
Conclusions
TP53 was the most common mutant driver gene accompanying EGFR mutation in stage IIb and IIIa resected NSCLC, followed by NOTCH1 mutations. Mutations of ATM, KIT, FGFR2, MET, PDGFRA, RB1, and wildtype NOTCH1, ERBB4, FGFR3 were independent risk factors for the recurrence of resected EGFR mutant NSCLC.
Acknowledgments
We want to thank the participants and their relatives for their cooperation and contribution to this work. The authors thank Shanghai Tongshu Biotechnology Co., Ltd. for technical support.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was carried out in compliance with the Declaration of Helsinki Principles and was approved by the Ethics Committee of the First Affiliated Hospital of Guangzhou Medical University (No. CCTC1501).
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299-311. [Crossref] [PubMed]
- NSCLC Meta-analyses Collaborative Group, Arriagada R, Auperin A, et al. Adjuvant chemotherapy, with or without postoperative radiotherapy, in operable non-small-cell lung cancer: two meta-analyses of individual patient data. Lancet 2010;375:1267-77. [Crossref] [PubMed]
- He Q, Xin P, Zhang M, et al. The impact of epidermal growth factor receptor mutations on the prognosis of resected non-small cell lung cancer: a meta-analysis of literatures. Transl Lung Cancer Res 2019;8:124-34. [Crossref] [PubMed]
- Nagasaka M, Gadgeel SM. Role of chemotherapy and targeted therapy in early-stage non-small cell lung cancer. Expert Rev Anticancer Ther 2018;18:63-70. [Crossref] [PubMed]
- Lv C, Ma Y, Feng Q, et al. A pilot study: sequential gemcitabine/cisplatin and icotinib as induction therapy for stage IIB to IIIA non-small-cell lung adenocarcinoma. World J Surg Oncol 2013;11:96. [Crossref] [PubMed]
- Zito Marino F, Ronchi A, Accardo M, et al. Concomitant ALK/KRAS and ALK/EGFR mutations in non small cell lung cancer: different profile of response to target therapies. Transl Cancer Res 2017;6:S457-60. [Crossref]
- Zhou JX, Yang H, Deng Q, et al. Oncogenic driver mutations in patients with non-small-cell lung cancer at various clinical stages. Ann Oncol 2013;24:1319-25. [Crossref] [PubMed]
- Jao K, Tomasini P, Kamel-Reid S, et al. The prognostic effect of single and multiple cancer-related somatic mutations in resected non-small-cell lung cancer. Lung Cancer 2018;123:22-9. [Crossref] [PubMed]
- Hu W, Liu Y, Chen J. Concurrent gene alterations with EGFR mutation and treatment efficacy of EGFR-TKIs in Chinese patients with non-small cell lung cancer. Oncotarget 2017;8:25046-54. [PubMed]
- Chen M, Xu Y, Zhao J, et al. Concurrent Driver Gene Mutations as Negative Predictive Factors in Epidermal Growth Factor Receptor-Positive Non-Small Cell Lung Cancer. EBioMedicine 2019;42:304-10. [Crossref] [PubMed]
- Zhao ZR, Lin YB, Ng CSH, et al. Mutation Profile of Resected EGFR-Mutated Lung Adenocarcinoma by Next-Generation Sequencing. Oncologist 2019;24:1368-74. [Crossref] [PubMed]
- Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): amulticentre, open- label, randomised, phase 3 study. Lancet Oncol 2011;12:735-42. [Crossref] [PubMed]
- Yan X, Wang H, Li P, et al. Efficacy of first-line treatment with epidermal growth factor receptor-tyrosine kinase inhibitor (EGFR-TKI) alone or in combination with chemotherapy for advanced non-small cell lung cancer (NSCLC) with low-abundance mutation. Lung Cancer 2019;128:6-12. [Crossref] [PubMed]
- Blakely CM, Watkins TBK, Wu W, et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet 2017;49:1693-704. [Crossref] [PubMed]
- Sanchez-Vega F, Mina M, Armenia J, et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018;173:321-37.e10. [Crossref] [PubMed]
- Nowell CS, Radtke F. Notch as a tumour suppressor. Nat Rev Cancer 2017;17:145-59. [Crossref] [PubMed]
- Xie M, Zhang L, He CS, et al. Activation of Notch-1 enhances epithelial-mesenchymal transition in gefitinib-acquired resistant lung cancer cells. J Cell Biochem 2012;113:1501-13. [PubMed]
- Xie M, He CS, Wei SH, et al. Notch-1 contributes to epidermal growth factor receptor tyrosine kinase inhibitor acquired resistance in non-small cell lung cancer in vitro and in vivo. Eur J Cancer 2013;49:3559-72. [Crossref] [PubMed]
- Ware KE, Marshall ME, Heasley LR, et al. Rapidly acquired resistance to EGFR tyrosine kinase inhibitors in NSCLC cell lines through de-repression of FGFR2 and FGFR3 expression. PLoS One 2010;5:e14117. [Crossref] [PubMed]
- Behrens C, Lin HY, Lee JJ, et al. Immunohistochemical expression of basic fibroblast growth factor and fibroblast growth factor receptors 1 and 2 in the pathogenesis of lung cancer. Clin Cancer Res 2008;14:6014-22. [Crossref] [PubMed]
- Cecchi F, Rabe DC, Bottaro DP. Targeting the HGF/Met signalling pathway in cancer. Eur J Cancer 2010;46:1260-70. [Crossref] [PubMed]
- Lu HY, Zhang G, Cheng QY, et al. Expression and mutation of the c-KIT gene and correlation with prognosis of small cell lung cancer. Oncol Lett 2012;4:89-93. [Crossref] [PubMed]
- Xiao H, Wang J, Liu Y, et al. Relative influence of c-Kit expression and epidermal growth factor receptor gene amplification on survival in patients with non-small cell lung cancer. Oncol Lett 2014;8:582-8. [Crossref] [PubMed]
- Helman E, Nguyen M, Karlovich CA, et al. Cell-Free DNA Next-Generation Sequencing Prediction of Response and Resistance to Third-Generation EGFR Inhibitor. Clin Lung Cancer 2018;19:518-30.e7. [Crossref] [PubMed]
- Offin M, Chan JM, Tenet M, et al. Concurrent RB1 and TP53 alterations define a subset of EGFR-mutant lung cancers at risk for histologic transformation and inferior clinical outcomes. J Thorac Oncol 2019;14:1784-93. [Crossref] [PubMed]
- Shen P, Zhong W. Adjuvant EGFR TKI therapy for resectable non-small cell lung cancer: new era for personalized medicine. J Thorac Dis 2018;10:1364-69. [Crossref] [PubMed]