
EGFR mutation genotypes affect efficacy and resistance mechanisms of osimertinib in T790M-positive NSCLC patients
Introduction
Epidermal growth factor receptor (EGFR) mutations are found in almost half of Asian non-small cell lung cancer (NSCLC) patients (1,2). Treatment with EGFR tyrosine kinase inhibitors (TKIs) is a milestone achievement in the management of advanced EGFR-mutant NSCLC. However, it is also of note that increasing evidence indicates EGFR-mutant NSCLCs display broad molecular and clinical heterogeneity (3-11). Even the most two common EGFR TKI-activating mutations, exon 19 deletion (19Del) and L858R, have differences in sensitivities to first- and second-generation EGFR TKIs (e.g., gefitinib, erlotinib, and afatinib) (4-6). In addition, these two types of EGFR mutation also produced differences in resistance mechanisms to first- or second-generation TKIs, where the acquired T790M mutation occurred more frequently in the 19Del mutant tumors than in L858R mutant tumors (8,9). Recent studies still revealed that distinct EGFR-mutant tumors have differences in tumor mutation burden (TMB) and in outcomes with immune checkpoint blockade treatment, where 19Del mutant tumors have a significantly lower TMB and a reduced benefit of treatment with immune checkpoint inhibitors compared with L858R mutant tumors (10,11).
Moreover, in contrast to L858R mutation, 19Del mutations have a variety of variants and represent a more heterogeneous disease entity (12,13). Distinct 19Del variants also confer heterogeneous sensitivity to first- or second-generation EGFR TKIs (14-19), and even associate with different histology in NSCLCs (19). Additional studies still suggested that patients carrying the canonical variant p.E746_A750del have a higher proportion of acquired T790M mutation than those carrying other 19Del variants after progression on earlier-generation TKIs (20,21).
Osimertinib is a potent third-generation EGFR TKI with robust activity in advanced EGFR-mutant NSCLCs, including those with T790M resistance mutation (22). Although treatment responses and acquired resistance mechanisms to earlier-generation EGFR TKIs between EGFR mutation types or subtypes have been identified to be different in previous studies, few have comprehensively examined whether there is a discrepancy in sensitivity and resistance mechanisms to osimertinib between EGFR-mutant genotypes in the presence of T790M mutation. This study was thus performed with the purposes of assessing the differences in clinical outcomes and resistance mechanisms among patients with T790M-positive NSCLCs who were specifically receiving osimertinib treatment and harboring different EGFR mutation types and subtypes.
Methods
Study population
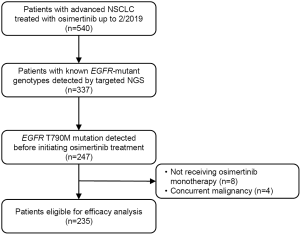
Following the institutional review board approval at Sun Yat-sen University Cancer Center (SYSUCC), we identified all patients with advanced NSCLC treated with osimertinib up to February 2019 in our institutional lung cancer-specific database. Patients were included if they had a detection of T790M mutation before commencing osimertinib treatment, and a known EGFR-mutant genotype determined by next-generation sequencing (NGS). Patients were excluded if they received osimertinib as maintenance treatment or in combination with chemotherapy or other targeted therapy, as well as those with concurrent malignancy (Figure S1).
Data collection
Patient clinicopathologic features and treatment histories were retrospectively collected from electronic medical records. Clinical outcomes include overall response rate (ORR), clinical benefit rate (CBR), progression-free survival (PFS), and overall survival (OS). To determine the response, radiographic scans were reviewed by investigators (QZ and WF) based on Response Evaluation Criteria in Solid Tumors version 1.1 (23). ORR was defined as the proportion of patients with a best overall response of complete response (CR) or partial response (PR). CBR was defined as the proportion of patients whose best overall response is either CR, PR, or stable disease lasting for more than 24 weeks. PFS was defined as the date of commencing osimertinib treatment to the date of disease progression or death, whichever occurred first. Patients who were alive without disease progression were censored on the date of their last efficacy assessment. OS was defined as the date of commencing osimertinib treatment to death. Patients who were still alive were censored at the last contact date.
Identification of EGFR genotypes
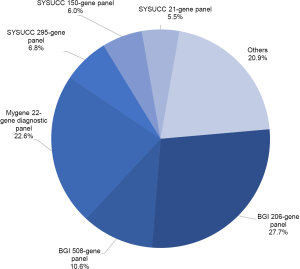
EGFR genotypes were dominantly determined by using one of the following commercial NGS panels: the BGI Oseq-Drug panel, the Mygene Diagnostic panel, and the Personalized Diagnostics NGS panels of our hospital. During our study, the BGI Oseq-Drug panel expanded from 206 genes to a 508-gene panel. The Mygene Diagnostic panel was used to sequence targeted hotspots for 22 lung cancer-related genes. The SYSUCC Personalized Diagnostics NGS panels include a 21-gene panel for circulating tumor DNA (ctDNA), a 150-gene ctDNA panel, and a 295-gene panel for solid tumor, and all panels have full coverage of EGFR gene. In addition, a subset of patients was sequenced using other targeted NGS panels at outside institutions (Figure S2).
Osimertinib resistance mechanism analysis
To evaluate the osimertinib resistance mechanisms among different EGFR genotypes, we further identified patients who conducted NGS testing at osimertinib resistance. Either tumor tissue or plasma cfDNA analysis was available. We focused on two common established mechanisms, the EGFR T790M loss and the C797S acquisition. Methodologies to detect EGFR resistance mutations included the BGI Oseq-Drug panel, the Mygene Diagnostic panel, and the SYSUCC Personalized Diagnostics NGS panels. In addition, a public cohort of 93 lung cancer patients with osimertinib-resistance mutation profiles from Yang et al. (24) was also used to evaluate osimertinib resistance mechanisms according to EGFR genotypes. The patients in public cohort were enrolled from multiple centers across China, and their mutation profiles were tested using a targeted NGS panel for 416 cancer-related genes. Moreover, we also identified patients who underwent tumor biopsy at osimertinib resistance. These patients were analyzed for the resistance mechanism of small-cell lung cancer (SCLC) transformation. All of their pathology slides were examined by expert lung cancer pathologists.
Statistical analysis
Differences in baseline characteristics between patients with EGFR mutation types or subtypes were compared using chi-square or Fisher’s exact test for categorical data and the Wilcoxon rank-sum test for continuous data. PFS and OS were assessed by the Kaplan-Meier curve with log-rank test. Differences in tumor responses and EGFR resistance mechanisms were compared between patient groups by using chi-square tests. Hazard ratios (HRs) and corresponding 95% confidence intervals (CIs) were calculated by using univariate and multivariate Cox proportional hazards regression models to assess the effects of different variables on PFS and OS. Statistical analyses were performed using STATA/MP 14.0 and GraphPad Prism software, version 7.0. Two-tailed P<0.05 was considered statistically significant.
Results
Characteristics of patients receiving osimertinib
We identified 540 advanced NSCLC patients treated with osimertinib between August 2015 and February 2019. From 337 patients with known mutant EGFR genotyping results determined by NGS, 90 had no detection of T790M mutation, 8 received a combination therapy, and 4 had concurrent malignant neoplasm. Finally, 235 patients were included in association analysis between EGFR genotypes and clinical outcomes of osimertinib treatment. Patient characteristics are summarized in Table 1. Among 235 patients [median age, 55 years (range, 27–81 years); 143 (60.9%) female; 164 (69.8%) non-smokers; 227 (96.6%) adenocarcinoma], 136 (57.9%) harbored EGFR 19Del, 93 (39.6%) harbored L858R, and only 6 (2.6%) harbored uncommon EGFR mutations (two G719A, one G719A+L861Q, one G719A+E709A, one V769M, and one exon 20 insertion [p.A763_Y764insFQEA]). Patient characteristics grouped by EGFR mutation types are provided in Table S1. For T790M mutation status at baseline prior to osimertinib treatment, 101 (43.0%) patients were detected using plasma specimens, 98 (41.7%) using tumor tissue specimens, 8 (3.4%) using both ctDNA and tissue specimens, 15 (6.4%) using pleural/peritoneal effusion specimens, 2 (0.9%) using cerebrospinal fluid specimens, and 11 (4.7%) were unspecified.

Full table
Full table
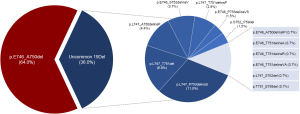
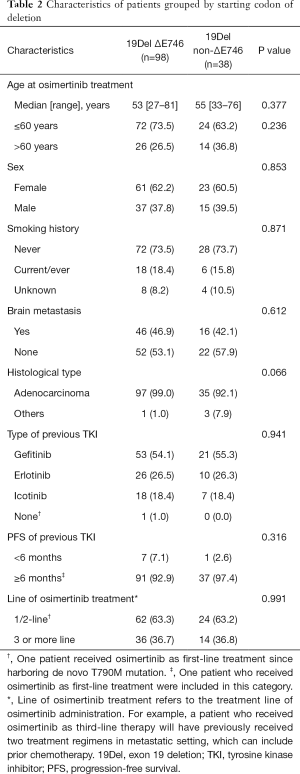
In addition, we identified 14 deletion variants from the 136 patients with EGFR 19Del (Figure S3). The most common deletion variants were p.E746_A750del (n=87, 64.0%), followed by p.L747_P753delinsS (n=15, 11.0%), p.L747_T751del (n=9, 7.1%), and p.L747_A750delinsP (n=6, 4.4%), consistent with previously reported frequencies for patients harboring 19Del (14-19). We classified these patients into two subgroups according to the deletion starting codon by Gazdar et al. (13): from E746 [ΔE746 subgroup, n=98 (72.1%)] or from other than E746 [non-ΔE746 subgroup, n=38 (27.9%)]. In non-ΔE746 subgroup, 35 patients harbored deletions from codon L747, two S752, and one T751. There were no clinicopathologic differences between two subgroups at baseline, except for a trend toward more non-adenocarcinoma in non-ΔE746 versus ΔE746 patients (7.9% vs. 1.0%; P=0.066; Table 2).
Full table
Distinct EGFR mutation types have similar outcomes with osimertinib treatment
At the time of analysis (data cutoff September 1, 2019), the median follow-up was 18.0 months. Overall, 233 patients were evaluable for response. The ORR and CBR to osimertinib in our cohort were 53.7% and 78.1%, respectively. Besides, 169 patients either had disease progression or had died at the time of data cutoff. The median PFS of overall population was 10.1 months (95% CI: 9.1–11.5 months; Figure S4A), and the median OS was 22.0 months (95% CI: 18.7–24.4 months; Figure S4B). We examined the effect of distinct EGFR mutation types on outcome with osimertinib treatment. We found that patients harboring 19Del, L858R, and uncommon mutations had non-significantly different ORRs (58.5%, 47.3%, and 40.0%, respectively, P=0.206, Figure S5A) and CBRs (81.5%, 73.1%, and 80%, respectively, P=0.322, Figure S5B). There were also no significant difference in PFSs (median: 11.3, 9.0, and 11.7 months, respectively, log-rank P=0.276, Figure S5C) and OSs (median: 22.6, 20.1 months, and not reached, respectively, log-rank P=0.173, Figure S5D) among patients with 19Del, L858R, and uncommon mutations.
Distinct EGFR 19Del subtypes have different outcomes with osimertinib treatment
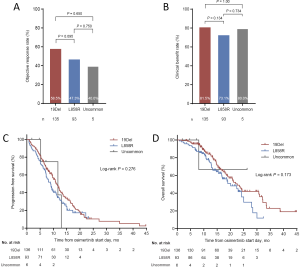
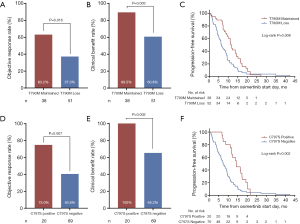
Due to the absence of study reporting the association between EGFR 19Del subtypes and osimertinib efficacy, we subsequently investigated the impact of varying EGFR 19Del subtypes on clinical outcomes of osimertinib treatment in our cohort of 136 T790M-mutant cases. We compared these cases with patients with L858R tumors. We found ΔE746 tumors had a significantly higher ORR and CBR compared with L858R tumors (ORR: 64.9% vs. 47.3%, P=0.014; CBR: 86.6% vs. 73.1%, respectively, P=0.020), whereas non-ΔE746 tumors had similar ORR and CBR compared with L858R tumors (ORR: 42.1% vs. 47.3%, respectively, P=0.587; CBR: 68.4% vs. 73.1%, respectively, P=0.588) (Figure 1A,B). PFS and OS in ΔE746 patients were also significantly longer (median PFS: 12.2 vs. 9.0 months, log-rank P=0.027; median OS: 23.7 vs. 20.1 months, log-rank P=0.02) whereas non-ΔE746 patients had similar PFS and OS compared with the L858R patients (median PFS: 8.6 vs. 9.0 months, log-rank P=0.621; median OS: 15.7 vs. 20.1 months, log-rank P=0.955) (Figure 1C,D). Moreover, with regard to ΔE746 subgroup, no significant differences in PFS and OS were observed between the most common 19Del variant p.E746_A750del and other ΔE746 variants (log-rank P=0.967, P=0.396, respectively; Figure S6A,B). Overall, these data suggested that patients with EGFR ΔE746 tumors have a significantly better benefit from osimertinib treatment.

Clinicopathologic features associated with osimertinib outcomes
We further evaluated the effect of clinical and pathologic features on PFS and OS to osimertinib in patients with T790M-mutant NSCLC. In univariate analysis, we found that tumor histological type, line of osimertinib therapy, and EGFR mutation subtype were significant prognostic factors for PFS (Table S2). In multivariate Cox regression analysis adjusting for potential confounders, only tumor histologic type and EGFR mutation subtype remained independent predictive value for PFS (Table S2). Specifically, EGFR non-ΔE746 (HR=1.70, 95% CI: 1.08–2.69, P=0.022) and L858R (HR=1.70, 95% CI: 1.19–2.43, P=0.004) had a significantly higher HR than ΔE746 subtype. Similarly, multivariate analysis also confirmed that L858R (HR=1.70, 95% CI: 1.06–2.73, P=0.026) was independently associated with worse OS than EGFR ΔE746 mutation subtype (Table S3). In addition, non-adenocarcinoma histology (HR=3.92, 95% CI: 1.49–10.27, P=0.006) and third- or more line of osimertinib therapy (HR=1.59, 95% CI: 1.03–2.46, P=0.035) were also independently associated with worse OS for osimertinib treatment.
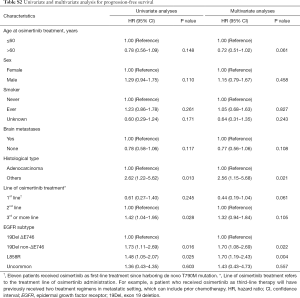
Full table
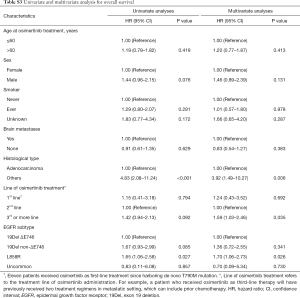
Full table
Distinct EGFR genotypes have different resistance mechanisms to osimertinib treatment
Given the reported association between mechanisms of osimertinib resistance and duration of osimertinib treatment (25-27), we investigated the osimertinib resistance mechanisms across variable EGFR mutation subtypes in our cohort. Of 169 patients developed resistance to osimertinib, we identified 90 patients who underwent NGS testing after resistance to osimertinib, including 34 (37.8%) using tissue biopsy specimens, 46 (51.1%) using plasma specimens, 7 (7.8%) using pleural effusion specimens, and 3 (3.3%) using both tissue and plasma specimens. Across all EGFR mutation subtypes, T790M loss was seen in 52 patients (57.8%); while acquired C797S mutation was developed in 20 patients (22.2%), all with maintained T790M. Of these C797S mutations, 15 (75.0%) were in cis to T790M, 2 (10.0%) were in trans, but 3 (15.0%) were not specified. As expected, patients with T790M loss had a significantly lower ORR (37.3% vs. 63.2%, P=0.016; Figure 2A), lower CBR (60.8% vs. 89.5%, P=0.003; Figure 2B), and shorter duration of osimertinib treatment (6.4 vs. 11.3 months, Log-rank P=0.008; Figure 2C) compared with patients with T790M maintained. Patients with acquired C797S mutation had a significantly higher ORR (75.0% vs. 40.6%, P=0.007; Figure 2D), higher CBR (100% vs. 65.2%, P=0.002; Figure 2E), and longer duration of osimertinib treatment (14.2 vs. 7.3 months, Log-rank P=0.002; Figure 2F) compared with patients without acquired C797S mutation.
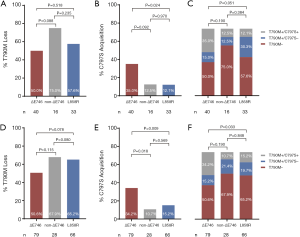
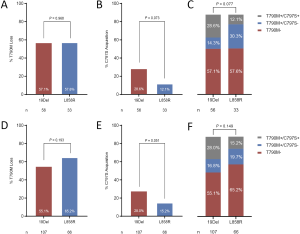
Comparison of T790M loss between patients with 19Del versus L858R did not reveal any significant differences (32 of 56 patients, 57.1% vs. 19 of 33 patients, 57.6%, P=0.968, Figure S7A). Both EGFR ΔE746 and non-ΔE746 tumors had equivalent rate of T790M loss compared with L858R tumors (20 of 40 patients, 50.0% vs. 19 of 33 patients, 57.6%, P=0.518; 12 of 16 patients, 75.0% vs. 19 of 33 patients, 57.6%, P=0.235; respectively, Figure 3A). However, it is noteworthy that borderline statistical significance was observed when comparing difference in acquired C797S mutation between 19Del and L858R mutation types (16 of 56 patients, 28.6% vs. 4 of 33 patients, 12.1%, P=0.073, Figure S7B). Moreover, ΔE746 tumors had a significantly higher (14 of 40 patients, 35.0% vs. 4 of 33 patients, 12.1%, P=0.024; Figure 3B) whereas non-ΔE746 tumors had a similar (2 of 16 patients, 12.5% vs. 4 of 33 patients, 12.1%, P=0.970; Figure 3B) prevalence of developing acquired C797S mutation compared with L858R tumors. According to the T790M and C797S mutation status, we classified the resistance mechanisms into three types including T790M loss, T790M maintained without C797S acquisition, and T790M maintained with C797S acquisition. Comparison of these resistance mechanisms between patients with 19Del versus L858R revealed a borderline statistical difference (P=0.077, Figure S7C). Further comparisons between ΔE746 and non-ΔE746 versus L858R tumors suggested that resistance pattern of ΔE746 was different from L858R (P=0.051, Figure 3C), while non-ΔE746 was similar (P=0.384, Figure 3C). A combined analyses of our institutional cohort and the public multicenter cohort (24) confirmed no differences in T790M loss between 19Del versus L858R tumors (59 of 107 patients, 55.1% vs. 43 of 66 patients, 65.2%, P=0.193, Figure S7D), or between ΔE746 and non-ΔE746 versus L858R tumors (40 of 79 patients, 50.6% vs. 43 of 66 patients, 65.2%, P=0.078; 19 of 28 patients, 67.9% vs. 43 of 66 patients, 65.2%, P=0.80; respectively, Figure 3D). In addition, a strong tendency towards statistical difference was found in acquired C797S mutation between 19Del and L858R mutation types (30 of 107 patients, 28.0% vs. 10 of 66 patients, 15.2%, P=0.051, Figure S7E). Moreover, combined analyses also confirmed ΔE746 tumors had a significantly higher frequency of acquired C797S mutation than non-ΔE746 (27 of 79 patients, 34.2% vs. 3 of 28 patients, 10.7%, P=0.018, Figure 3E) and L858R tumors (27 of 79 patients, 34.2% vs. 10 of 66 patients, 15.2%, P=0.009, Figure 3E). Resistance pattern was not different between 19Del and L858R tumors (P=0.149, Figure S7F), as well as between non-ΔE746 and L858R (P=0.848, Figure 3F), but was significantly different between ΔE746 and L858R (P=0.033, Figure 3F).
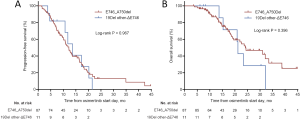
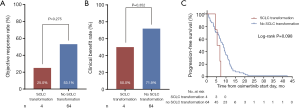
Of the 169 osimertinib-resistant patients, 71 underwent a post-progression tumor biopsy, and 68 (95.8%) of them succeeded to obtain tumor tissue. Four (5.9%) of 68 patients developed transformation to small-cell lung cancer (SCLC). All of them had 19Del mutation types (three ΔE746 and one non-ΔE746). However, statistical significance was not reached when compared with L858R tumors (4 of 45, 8.9% vs. 0 of 22, 0%, respectively, P=0.294). In addition, ORR, CBR, and duration of osimertinib treatment were all not significantly different between patients who developed SCLC transformation and those not developed SCLC transformation (Figure 4).
Discussion
In this study, we did not identify T790M-positive NSCLC patients carrying EGFR 19Del mutations had a significantly better therapeutic response or survival benefit to osimertinib compared with those carrying L858R or uncommon mutations. However, we did observe a subset of 19Del mutant patients (ΔE746) had superior clinical outcomes with osimertinib treatment than L858R mutant patients. Our study also found ΔE746 tumors had a higher whereas non-ΔE746 tumors had a similar proportion of acquired C797S mutation at resistance to osimertinib compared with L858R tumors. To the best of our knowledge, this is the first study to date to investigate the clinical effect of 19Del variants on outcomes of osimertinib treatment in T790M-positive NSCLCs, and the first analysis to compare osimertinib resistance mechanisms according to 19Del variants.
Indeed, several key studies have suggested that, in the presence of acquired T790M mutation, patients harboring EGFR 19Del mutations have a non-significantly higher response and longer PFS than those harboring L858R when receiving osimertinib treatment (22,28,29). Our analyses also observed this association in patients with EGFR T790M mutation treated with osimertinib. In contrast to these findings, however, there were some studies showed significant differences in PFS and OS between these two EGFR mutation types (30,31). These conflicting results may be due partly to differences in clinicopathologic characteristics and previous treatment histories among these studies. In our study, we observed that none but one patient received afatinib before osimertinib treatment. This was very different from other studies (22,28-31). We think that it may be associated with the facts that afatinib provides only a modest improvement in PFS but has higher rate of serious drug-related adverse events than gefitinib (32,33), and has not been covered by the basic medical insurance system in China before October, 2018.
In addition, it is known that EGFR 19Del mutants constitute a heterogeneous group of genetic alterations characterized by in-frame deletions, substitutions and insertions. In our study, we did observe 19Del ΔE746 tumors had higher sensitivity to osimertinib and more likelihood of developing acquired C797S mutation at resistance than L858R tumors, but non-ΔE746 tumors did not, which has never been previously reported. The underlying causes responsible for these discrepancies are still unclear, though it may be related to the variable sample sizes across different subgroups. The outcomes of osimertinib treatment were similar to those of first- and second-generation EGFR TKIs, where EGFR ΔE746 tumors are found to be associated with longer treatment duration of earlier-generation TKIs compared with non-ΔE746 tumors in several studies (17,18), highlighting potential differences between EGFR 19Del subtypes. The inconsistent findings of the effect of 19Del and L858R mutations on osimertinib efficacy in different studies may also due in part to different proportion of patients with various 19Del subtypes between studies (22,28-31). In fact, in vitro cellular experiments have observed that distinct 19Del variants confer different sensitivities to EGFR TKIs, including osimertinib (15,34,35). In addition, one recent study using molecular dynamics simulation analysis has also identified that structural consequences of p.L747_A750delinsP mutation, one common non-ΔE746 variant, reduces the sensitivity of osimertinib by impairing drug binding (15). L747 was a pivotal hydrophobic core stabilizing the inactive form of EGFR, changing in this site may disrupt the hydrophobic core and favored the active conformation of EGFR (36). Although these studies were not performed in the context of the T790M mutation, structural change in the deletion mutants may lead to different osimertinib sensitivity between ΔE746 and non-ΔE746 tumors. However, further prospective studies with larger sample sizes to identify the impact of EGFR genotypes on outcomes with osimertinib treatment are warranted, as well as more in-depth fundamental researches to explore the intrinsic mechanisms.
The emergence of EGFR T790M in 19Del tumor is higher than that in L858R tumors after earlier-generation EGFR TKIs targeted therapy (8,9). This may be the result of higher sensitivity and thus longer exposure to EGFR TKIs in 19Del tumors than in L858R tumors. Similarly, T790M-positive EGFR mutant tumors which gain more durable benefit from osimertinib targeted therapy could be more dependent on EGFR pathway. This may be necessitated for the development of on-target acquired resistance (C797S) to osimertinib. Our study results also supported this notion, where ΔE746 tumors have higher sensitivity to osimertinib and consequently have more likelihood of developing on-target acquired resistance to osimertinib. Moreover, patients with acquired C797S mutation did have better response and survival, with a CBR of 100%, and a higher ORR and longer treatment duration of osimertinib than patients without developing acquired C797S mutation.
Recent reports have also identified EGFR G724S as an emerging mutation mediating acquired resistance to osimertinib (37-41). Moreover, G724S as an allele-specific osimertinib-resistance mutation exists exclusively in the context of specific 19Del mutants but not the most common 19Del variant p.E746_A750del or L858R (41), supporting that potentially different mechanisms confer resistance to different mutant tumors. Our study results also identified two patients with G724S resistant mutations, both with original EGFR genotype of p.E746_S752delinsV. This incidence was similar to most previous reports (37-39), except for the study from Fassunke et al. (40). However, because of the relatively low occurrence frequency, no separate comparison was performed between patient groups in our study.
Some limitations of present study should be concerned. First, this retrospective, single-center study was still limited in the number of patients with rare mutations, as well as the total number of rare 19Del subtypes, which may not be representative of all NSCLC patients with EGFR rare mutations. Further large, prospective, multicentre studies are warranted to confirm and expand on our findings. Second, the NGS testing in this study was not performed using a single central panel, and not all resistant patients had performed post-osimertinib NGS testing and rebiopsy. However, we deemed that the various panels used in our study would not be limited in their ability of detecting specific EGFR 19Del variants or resistance mutations such as T790M and C797S. Additionally, other specific resistance mechanisms of osimertinib treatment, such as c-MET amplification, HER2 amplification, and BRAF mutations, etc. (42), were not assessed in our analysis. Moreover, osimertinib has been recently approved as first-line treatment before acquiring T790M mutation (43). Whether different sensitivity to osimertinib continuing to be observed in first-line setting remains unanswered.
Conclusions
Our study provided evidence supporting that not all EGFR 19Del tumors have equally favorable outcomes to osimertinib treatment in the context of the T790M mutation, where ΔE746 tumors suggest preferable sensitivity to osimertinib. Our analyses also revealed ΔE746 tumors have a higher frequency to develop the acquired C797S mutation after osimertinib treatment, highlighting potential different resistance mechanisms between EGFR 19Del variants. Genotyping of 19Del mutations should be precisely identified at diagnosis in order to determine the prognostic and predictive value and select the better treatment. Further researches are needed to determine the intrinsic mechanisms of our findings.
Acknowledgments
Funding: This work was supported by National Key R&D Program of China (Grant numbers 2016YFC0905500, 2016YFC0905503), Science and Technology Program of Guangdong (Grant numbers 2017B020227001, 2016A020215084), Science and Technology Program of Guangzhou (Grant number 201607020031), Chinese National Natural Science Foundation Project (Grant numbers 81772476, 81602005, 81872499), Pearl River Nova Program of Guangzhou (Grant number 201610010048), Outstanding Young Talents Program of Sun Yat-sen University Cancer Center (Grant number 16zxyc04) and Central Basic Scientific Research Fund for Colleges-Young Teacher Training Program of Sun Yat-sen University (Grant number 17ykpy81).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr.2020.03.35). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Institutional Review Board of Sun Yat-sen University Cancer Center (B2019-127-01), and was in accordance with the guidelines of the Helsinki Declaration (as revised in 2013) and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Shi Y, Au JS, Thongprasert S, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol 2014;9:154-62. [Crossref] [PubMed]
- Han B, Tjulandin S, Hagiwara K, et al. EGFR mutation prevalence in Asia-Pacific and Russian patients with advanced NSCLC of adenocarcinoma and non-adenocarcinoma histology: The IGNITE study. Lung Cancer 2017;113:37-44. [Crossref] [PubMed]
- Carey KD, Garton AJ, Romero MS, et al. Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer Res 2006;66:8163-71. [Crossref] [PubMed]
- Zhang Y, Sheng J, Kang S, et al. Patients with exon 19 deletion were associated with longer progression-free survival compared to those with L858R mutation after first-line EGFR-TKIs for advanced non-small cell lung cancer: a meta-analysis. Plos One 2014;9:e107161. [Crossref] [PubMed]
- Lee CK, Wu YL, Ding PN, et al. Impact of Specific Epidermal Growth Factor Receptor (EGFR) Mutations and Clinical Characteristics on Outcomes After Treatment With EGFR Tyrosine Kinase Inhibitors Versus Chemotherapy in EGFR-Mutant Lung Cancer: A Meta-Analysis. J Clin Oncol 2015;33:1958-65. [Crossref] [PubMed]
- Yang JC, Wu YL, Schuler M, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol 2015;16:141-51. [Crossref] [PubMed]
- Zhang Y, Chen G, Chen X, et al. The comparison of EGFR-TKI failure modes and subsequent management between exon 19 deletion and exon 21 L858R mutation in advanced non-small-cell lung cancer. J Cancer 2017;8:1865-71. [Crossref] [PubMed]
- Ke EE, Zhou Q, Zhang QY, et al. A Higher Proportion of the EGFR T790M Mutation May Contribute to the Better Survival of Patients with Exon 19 Deletions Compared with Those with L858R. J Thorac Oncol 2017;12:1368-75. [Crossref] [PubMed]
- Liang H, Pan Z, Wang W, et al. The alteration of T790M between 19 del and L858R in NSCLC in the course of EGFR-TKIs therapy: a literature-based pooled analysis. J Thorac Dis 2018;10:2311-20. [Crossref] [PubMed]
- Hastings K, Yu HA, Wei W, et al. EGFR mutation subtypes and response to immune checkpoint blockade treatment in non-small-cell lung cancer. Ann Oncol 2019;30:1311-20. [Crossref] [PubMed]
- Offin M, Rizvi H, Tenet M, et al. Tumor Mutation Burden and Efficacy of EGFR-Tyrosine Kinase Inhibitors in Patients with EGFR-Mutant Lung Cancers. Clin Cancer Res 2019;25:1063-9. [Crossref] [PubMed]
- Sharma SV, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 2007;7:169-81. [Crossref] [PubMed]
- Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene 2009;28 Suppl 1:S24-31. [Crossref] [PubMed]
- Rossi S, Toschi L, Finocchiaro G, et al. Impact of Exon 19 Deletion Subtypes in EGFR-Mutant Metastatic Non–Small-Cell Lung Cancer Treated With First-Line Tyrosine Kinase Inhibitors. Clin Lung Cancer 2019;20:82-7. [Crossref] [PubMed]
- Truini A, Starrett JH, Stewart TF, et al. The EGFR Exon 19 Mutant L747-A750>P Exhibits Distinct Sensitivity to Tyrosine Kinase Inhibitors in Lung Adenocarcinoma. Clin Cancer Res 2019;25:6382-91. [Crossref] [PubMed]
- Sutiman N, Tan SW, Tan EH, et al. EGFR Mutation Subtypes Influence Survival Outcomes following First-Line Gefitinib Therapy in Advanced Asian NSCLC Patients. J Thorac Oncol 2017;12:529-38. [Crossref] [PubMed]
- Kaneda T, Hata A, Tomioka H, et al. Possible differential EGFR-TKI efficacy among exon 19 deletional locations in EGFR-mutant non-small cell lung cancer. Lung Cancer 2014;86:213-8. [Crossref] [PubMed]
- Lee VHF, Tin VPC, Choy T, et al. Association of Exon 19 and 21 EGFR Mutation Patterns with Treatment Outcome after First-Line Tyrosine Kinase Inhibitor in Metastatic Non–Small-Cell Lung Cancer. J Thorac Oncol 2013;8:1148-55. [Crossref] [PubMed]
- Chung KP, Wu SG, Wu JY, et al. Clinical Outcomes in Non-Small Cell Lung Cancers Harboring Different Exon 19 Deletions in EGFR. Clin Cancer Res 2012;18:3470-7. [Crossref] [PubMed]
- Huang YH, Hsu KH, Tseng JS, et al. The Association of Acquired T790M Mutation with Clinical Characteristics after Resistance to First-Line Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor in Lung Adenocarcinoma. Cancer Res Treat 2018;50:1294-303. [Crossref] [PubMed]
- Zou B, Lee V, Chen L, et al. Deciphering mechanisms of acquired T790M mutation after EGFR inhibitors for NSCLC by computational simulations. Sci Rep 2017;7:6595. [Crossref] [PubMed]
- Yang JC, Ahn MJ, Kim DW, et al. Osimertinib in Pretreated T790M-Positive Advanced Non-Small-Cell Lung Cancer: AURA Study Phase II Extension Component. J Clin Oncol 2017;35:1288-96. [Crossref] [PubMed]
- Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228-47. [Crossref] [PubMed]
- Yang Z, Yang N, Ou Q, et al. Investigating Novel Resistance Mechanisms to Third-Generation EGFR Tyrosine Kinase Inhibitor Osimertinib in Non-Small Cell Lung Cancer Patients. Clin Cancer Res 2018;24:3097-107. [Crossref] [PubMed]
- Oxnard GR, Hu Y, Mileham KF, et al. Assessment of Resistance Mechanisms and Clinical Implications in Patients With EGFR T790M-Positive Lung Cancer and Acquired Resistance to Osimertinib. JAMA Oncol 2018;4:1527-34. [Crossref] [PubMed]
- Lin CC, Shih JY, Yu CJ, et al. Outcomes in patients with non-small-cell lung cancer and acquired Thr790Met mutation treated with osimertinib: a genomic study. Lancet Respir Med 2018;6:107-16. [Crossref] [PubMed]
- Zhao S, Li X, Zhao C, et al. Loss of T790M mutation is associated with early progression to osimertinib in Chinese patients with advanced NSCLC who are harboring EGFR T790M. Lung Cancer 2019;128:33-9. [Crossref] [PubMed]
- Jänne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015;372:1689-99. [Crossref] [PubMed]
- Goss G, Tsai CM, Shepherd FA, et al. Osimertinib for pretreated EGFR Thr790Met-positive advanced non-small-cell lung cancer (AURA2): a multicentre, open-label, single-arm, phase 2 study. Lancet Oncol 2016;17:1643-52. [Crossref] [PubMed]
- Igawa S, Ono T, Kasajima M, et al. Impact of EGFR genotype on the efficacy of osimertinib in EGFR tyrosine kinase inhibitor-resistant patients with non-small cell lung cancer: a prospective observational study. Cancer Manag Res 2019;11:4883-92. [Crossref] [PubMed]
- Auliac JB, Perol M, Planchard D, et al. Real-life efficacy of osimertinib in pretreated patients with advanced non-small cell lung cancer harboring EGFR T790M mutation. Lung Cancer 2019;127:96-102. [Crossref] [PubMed]
- Park K, Tan EH, O'Byrne K, et al. Afatinib versus gefitinib as first-line treatment of patients with EGFR mutation-positive non-small-cell lung cancer (LUX-Lung 7): a phase 2B, open-label, randomised controlled trial. Lancet Oncol 2016;17:577-89. [Crossref] [PubMed]
- Paz-Ares L, Tan EH, O'Byrne K, et al. Afatinib versus gefitinib in patients with EGFR mutation-positive advanced non-small-cell lung cancer: overall survival data from the phase IIb LUX-Lung 7 trial. Ann Oncol 2017;28:270-7. [Crossref] [PubMed]
- Furuyama K, Harada T, Iwama E, et al. Sensitivity and kinase activity of epidermal growth factor receptor (EGFR) exon 19 and others to EGFR-tyrosine kinase inhibitors. Cancer Sci 2013;104:584-9. [Crossref] [PubMed]
- Kohsaka S, Nagano M, Ueno T, et al. A method of high-throughput functional evaluation of EGFR gene variants of unknown significance in cancer. Sci Transl Med 2017;9. [PubMed]
- He M, Capelletti M, Nafa K, et al. EGFR exon 19 insertions: a new family of sensitizing EGFR mutations in lung adenocarcinoma. Clin Cancer Res 2012;18:1790-7. [Crossref] [PubMed]
- Oztan A, Fischer S, Schrock AB, et al. Emergence of EGFR G724S mutation in EGFR-mutant lung adenocarcinoma post progression on osimertinib. Lung Cancer 2017;111:84-7. [Crossref] [PubMed]
- Peled N, Roisman LC, Miron B, et al. Subclonal Therapy by Two EGFR TKIs Guided by Sequential Plasma Cell-free DNA in EGFR-Mutated Lung Cancer. J Thorac Oncol 2017;12:e81-4. [Crossref] [PubMed]
- Piotrowska Z, Isozaki H, Lennerz JK, et al. Landscape of Acquired Resistance to Osimertinib in EGFR-Mutant NSCLC and Clinical Validation of Combined EGFR and RET Inhibition with Osimertinib and BLU-667 for Acquired RET Fusion. Cancer Discov 2018;8:1529-39. [Crossref] [PubMed]
- Fassunke J, Muller F, Keul M, et al. Overcoming EGFR(G724S)-mediated osimertinib resistance through unique binding characteristics of second-generation EGFR inhibitors. Nat Commun 2018;9:4655. [Crossref] [PubMed]
- Brown BP, Zhang YK, Westover D, et al. On-target Resistance to the Mutant-Selective EGFR Inhibitor Osimertinib Can Develop in an Allele-Specific Manner Dependent on the Original EGFR-Activating Mutation. Clin Cancer Res 2019;25:3341-51. [Crossref] [PubMed]
- Ricordel C, Friboulet L, Facchinetti F, et al. Molecular mechanisms of acquired resistance to third-generation EGFR-TKIs in EGFR T790M-mutant lung cancer. Ann Oncol 2018;29:i28-37. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113-25. [Crossref] [PubMed]


