
Clinical impact of the lung tissue transcriptome in a teenager with multifocal invasive mucinous adenocarcinoma—a case report
Introduction
Lung cancer is the leading cause of death worldwide and the most common cancer, excluding skin cancers, after the prostate and breast cancers (1). The American Cancer Society’s estimates for 2019 about 228,150 new cases of lung cancer and 142,670 deaths in the United States with considerable Public Health impact.
The discovery of biomarkers for tumor prevention, early diagnosis and accurate prognosis, which involves a deeper understanding of the tumor histogenesis, would improve the management of lung cancer patient and “not yet patient” at risk to develop cancer.
To achieve this goal, genetic and molecular investigation must extend beyond the tumor specimen and include the histologically normal tissue adjacent to and distant from cancer.
Histologically normal-appearing tissues adjacent to cancers display cancer-associated molecular abnormalities known has “field effect”, that can either be due to “first hit” genetic alterations in a clone of cells from which the tumor ultimately develops, or to effects of the tumor on the local microenvironment (2-4). On the other hand, the prolonged exposure to genotoxic agents induces widespread molecular changes, defined as “field cancerization”, which can lead to the onset of multiple primary tumors (5). Decipher the transcriptional aberrations underlying these histologically normal-appearing conditions is proving to be fundamental in understanding the pathogenesis of tumor and, in particular, of the rare tumor histotypes in a singular clinical context, and in developing a personalized prognostic and therapeutic approach.
Invasive mucinous adenocarcinoma (IMA) is a distinct lung cancer histotype, which represents 5% of lung-adenocarcinomas, and mostly affects adults over 65 (6). Patients frequently present with pneumonia-like symptoms and multifocal disease (7). The prognostic implication is still a matter of debate, since some studies report a recurrence-free survival comparable to stage-matched non mucinous adenocarcinomas, while others describe a worse clinical behavior, with high recurrence rate (76% in 5 years) (8,9).
Here, we report the unprecedented case of a teenager with multifocal IMA and investigate the transcriptional signature of both the patient’s tumor and histologically normal lung tissues, distant from cancer. Integrated and comparative transcriptome analyses highlight the potential of the transcriptional profiles of normal and neoplastic lung tissues to unmask gene pathways driving oncogenesis with important clinical implications. We present the following case in accordance with the CARE guideline checklist.
Case presentation
A 16 years old girl was hospitalized due to acute adnexitis. Unexpectedly, the chest X-ray and subsequent spiral computed tomography (CT) imaging revealed, in the left-lower lobe, a nodule of 15 mm and, in the right middle lobe, a nodule of 7 mm with irregular margins (Figure 1A).
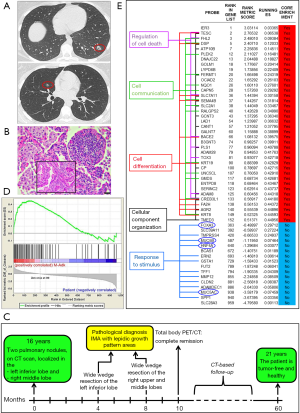
Signs and symptoms of lung cancer were absent. The girl had never smoked, but had a history of passive smoking in early life, and exposure to dichloroethylene and hexavalent chromium released by a tannery near her home. The paternal uncle died at the age of 46 due to metastatic lung cancer. No histopathological and molecular data available. The patient underwent a wide wedge resection of the left inferior lobe and lymph node sampling by video assisted thoracoscopic surgery. The pathological diagnosis was IMA with lepidic growth pattern areas and intratumoral lymphoid follicle-like structures (Figure 1B). The tumor stained negative for Transcription Termination Factor 1 (TTF-1), Caudal Type Homeobox 2 (CDX2), ALK Receptor Tyrosine Kinase (ALK) and ROS Proto-Oncogene 1 (ROS1). Molecular analyses revealed no Epidermal Growth Factor Receptor (EGFR) gene mutations, whereas a G12D mutation was found in the KRAS gene. Three months later, the smaller nodule was also removed by a wide wedge resection of the right upper and middle lobes, with lymph node sampling. Histopathological and molecular reports were consistent with previous findings. No therapy was given. Five years after initial diagnosis, a total body CT confirmed that the patient was tumor free and healthy (Figure 1C).
Histology and immunohistochemistry
The investigations were carried out following the rules of the Declaration of Helsinki of 1975, revised in 2013. The study was approved by the ethical committee of the “G. d’Annunzio” University (COET 19.07.2012) and written consent was obtained from the patient and parents.
For histology, lung tissue samples were formalin fixed and paraffin embedded, sectioned at 4 µm, and stained with hematoxylin and eosin (H&E). For immunohistochemistry, paraffin embedded sections, after antigen retrieval, were stained with the following antibodies (Abs), TTF-1 (clone 8G7G3/1, Agilent Technologies), CDX2 (clone DAK-CDX2, Agilent Technologies), ALK (clone D5F3, Roche), ROS1 (clone CST D4D6, Cell Signaling), C-X-C Motif Chemokine Ligand 13 (CXCL13; R&D Systems).
qBiomarker somatic mutation PCR Arrays
To assess KRAS and EGFR mutations, the DNA was extracted from 8 formalin fixed, paraffin embedded sections (10 µm). EGFR and KRAS mutations were analyzed with the qBiomarker™ Somatic Mutation PCR Arrays (Qiagen). Data were analysed with the Data analysis software for qBiomarker Somatic Mutation PCR Arrays (www.SABiosciences.com/somaticmutationdataanalysis.php).
Next generation sequencing (NGS)
RNA was extracted from fresh frozen neoplastic samples and histologically normal lung samples. According to The Cancer Genome Atlas (TCGA) Research Network protocols (http://cancergenome.nih.gov/), histologically normal tissues adjacent to tumor samples have been collected more than 2 cm distant from cancer, to avoid the field effect (10).
RNA was extracted from 10 frozen tissue sections (10 µm) and the NGS was performed by Lexogen GmbH, using the Illumina HiSeq 2500 V4 System. The reads alignment was done with STAR aligner and differential expression analysis was performed with DESeq software package.
Gene set enrichment analysis (GSEA)
For the GSEA, we used the GSEA computational method (http://software.broadinstitute.org/gsea/index.jsp), setting the IMA signature, identified by Guo et al. (11), as gene set.
Functional classification of differentially expressed genes
The functional classification of genes was performed using the Gene Ontology Consortium website (http://geneontology.org), with the PANTHER Overrepresentation Test (GO Ontology database, Released 2018-12-01), followed by Fisher’s Exact test with Bonferroni correction. Subsequently, the functional categories were also screened using the DAVID (12,13) and the UniProt database (https://www.uniprot.org/).
Comparative transcriptome analyses
The cohort of patients, identified by Guo et al. to determine the IMA signature, was used for the comparative transcriptome analyses with the IMA of the patient.
For comparative transcriptome analysis of normal lung tissues, the data of the reference cohort (consisting of women died for accidental causes and matched for age and smoking history) were obtained from the GTEx portal (https://gtexportal.org/home/) (V6p release).
The IMA signature is not enriched in the patient’s IMA
To assess whether, this unusual clinical case, was linked to a particular gene signature, we analyzed, through RNA-sequencing, the transcriptome of the patient’s IMA that was compared, through the GSEA method (14), to the recently defined IMA signature (11). The signature was not enriched in our patient (Figure 1D). Genes that regulate cell death, communication, differentiation and cellular component organization prevailed in IMA signature, whereas expression of genes regulating the response to stimulus, including typical IMA-associated genes Hepatocyte Nuclear Factor 4 Alpha (HNF4A), Forkhead Box A3 (FOXA3), Mucin 5AC (MUC5A) and Mucin 5B (MUC5B), was prominent in the patient’s IMA (Figure 1E).
The transcriptome of the patient’s normal lung reveals the down-regulation of genes driving cell adhesion and apoptosis and up-regulation of RAS family members and genes driving the response to chemical stimulus
Next, we wondered whether these alterations could also be found in the patient’s histologically normal lung tissue, far from cancer (10). Comparative transcriptome analyses with lung tissue of healthy age matched never smoker women revealed that genes driving the response to stimulus, chemical and organic substances, primarily CXCL8/IL-8, Atypical Chemokine Receptor 1 (ACKR1/DARC), and the member of the RAS oncogene family RAB7B were expressed in the patient’s normal lung, but absent in the lung of healthy women (Figure 2A).
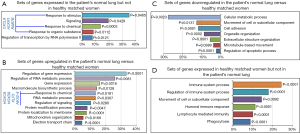
Additional genes driving the response to chemical, primarily Homeobox C9 (HOXC9), Homeobox D9 (HOXD9), Kruppel Like Factor 5 (KLF5), NK2 Homeobox 8 (NKX2-8), together with regulators of cell signaling, electron transport chain and mitochondrial organization, gene transcription and expression were upregulated in the patient’s normal lung (Figure 2B). Interestingly, genes driving cell adhesion, cell movement, metabolic and apoptotic processes were downregulated in patient’s normal lung (Figure 2C), whereas genes driving immune system processes, humoral and lymphocyte immune responses were expressed in healthy-woman’s lung, but lacked in the patient’s normal lung (Figure 2D).
The transcriptome of the patient’s IMA revealed overexpression of genes driving the immune system process and the response to chemical stimulus and down-regulation of specific proto-oncogenes
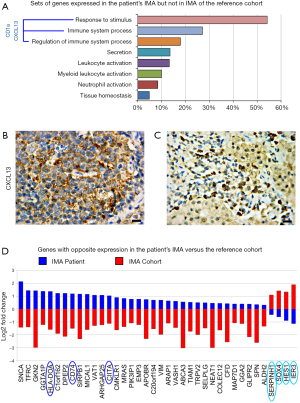
Comparative analyses, by functional categories, between patient’s IMA and IMA-signature cases revealed that genes driving the response to stimulus and immune system process, primarily CD1a and CXCL13/BCA-1, and genes involved in leukocyte, myeloid cell and neutrophil activation were expressed in the patient’s IMA, but lacked in the reference cohort (Figure 3A,B,C). Thirty seven genes revealed opposite expression. Specifically, in contrast to IMA signature cases, in the patient’s IMA 4/37 genes, Serpin Family H Member 1 (SERPINH1), SRY-Box 4 (SOX4), Hes Family BHLH Transcription Factor 1 (HES1) and Immediate Early Response 3 (IER3) were downmodulated, whereas 33/37 genes, including HLA-DOA, CD74 and Class II Major Histocompatibility Complex Transactivator (CIITA) were upregulated (Figure 3D).
Discussion
The present clinical case stands for the very young age of the patient who developed a rare histotype of lung cancer with multifocal onset and bilateral lung involvement.
According to the 8th TNM staging system of lung cancer, the matching of morphological, immunohistochemical and molecular profiles between the two neoplastic nodules was consistent with the diagnosis of intrapulmonary metastasis and classified the tumor as pathologic stage T1bN0M1a, i.e., stage IVA (15). However, the possibility of multiple primary cancers cannot be excluded (16).
Genes driving the response to stimulus, which include the typical IMA markers HNF4A, FOXA3, MUC5A and MUC5B (11,17), and the response to chemicals were overexpressed in both the patient’s tumor compared to the IMA signature, and the patient’s normal lung compared to control lungs. This molecular trait may have been triggered by long term exposure to passive smoking (18-20) and chemical carcinogens (21,22), although an inherited predisposition to early onset lung cancer cannot be ruled out (23).
Among the functional categories of genes driving the response to stimulus, chemical and organic substances, the genes coding for CXCL8/IL-8, ACKR1/DARC, and the member of the RAS oncogene family RAB7B were the most represented in the normal lung tissue of our young patient.
The multifunctional cytokine CXCL8/IL-8, by interacting with its cognate receptors, C-X-C chemokine receptor 1 (CXCR1) and CXCR2 mediates the initiation and development of various cancers, including lung cancer (24). Circulating CXCL8/IL-8 may predict the risk of lung cancer, since it is upregulated prior to clinical diagnosis (25,26). The Lung cancer screening trial has shown that high expression of CXCL8/IL-8 can raise the risk of lung cancer by 45–86% (25,26). CXCL8/CXCR1-2 axis promotes the transactivation of EGFR, which results in Ras GTPase activation and NSCLC cell proliferation (27).
ACKR1 binds over 20 inflammatory chemokines belonging to the CC and CXC families (28). It is expressed by erythrocytes and endothelial cells lining small veins and venules. Recent evidences suggest that ACKR1 expressed by erythrocytes acts as a chemokine buffer and can limit excessive leukocyte extravasation, whereas endothelial ACKR1 increases neutrophil and monocyte extravasation and promotes acute and chronic inflammation (29).
RAB7B is a lysosome-associated small GTPase, that regulates vesicular traffic from early to late endosomal stages of the endocytic pathway (30). It has been reported that increased expression of RAB7B, caused by growth factor withdrawal in mammalian cells, induces the downregulation of nutrient transporters, which activate stress response pathways and trigger apoptosis (31). However, its function and role in cancer remain to be defined.
Among the genes driving the response to chemicals, HOXC9, HOXD9, KLF5 and NKX2-8 were the most prevalent, and were upregulated in the patient’s normal lung when compared with age-matched healthy controls.
Both Homeobox transcription factors, HOXC9 and HOXD9, have been associated with increased risk to develop cancer and have been described as proto-oncogenes (32), but deciphering the cancer context specific role of these transcription factor remains a challenge.
KLF5 is a member of the zinc finger class of DNA-binding transcriptional regulators that regulates cell growth, proliferation, differentiation, apoptosis and tumorigenesis in a number of systems. It has been reported to act as a proto-oncogene in the lung, by promoting lung tumorigenesis via SOX4 up-regulation (33).
The function of NKX2-8 in lung cancer is still unclear. Indeed, knockout of NKX2-8 leads to proliferation of lung progenitor cells and widespread dysplasia in the large airways of mice (34), yet overexpression of this protein seems to enhance the tumorigenicity of malignant cell lines (35).
Co-activation of the TTF-1 and NKX2-8 pathways was discovered to confer resistance to cisplatin in lung cancer cell lines and was found to be associated with poor survival in NSCLC patients (36), however the meaning of NKX2-8 overexpression in the normal lung tissue is to be defined.
Notably, genes driving immune responses were found unexpressed in the patient's normal lung versus healthy control tissues, whereas genes governing cell adhesion, subcellular organization and apoptosis were dramatically downregulated, suggesting a favorable soil for the loss of cell-cell interactions and aberrant cell proliferation out of immunological control.
Molecular analyses of the neoplastic tissue revealed mutation of the KRAS gene, which is a common driver aberration in IMA (6) and occurs in our patient as KRAS codon 12 mutation (G12D). KRAS positive status significantly associates with IMA and MUC5B and MUC5AC expression (37). It has been demonstrated that a single endogenous mutant KRAS allele is sufficient to promote lung tumor formation, meanwhile malignant progression requires additional genetic alterations (38). KRAS G12D mutation, which induces a high-level of resistance to apoptosis and predisposition to anchorage-independent growth (39), may have synergized with the transcriptional alterations found in the patient’s normal lung tissue resulting in tumor progression. The immunomodulatory potential of KRAS mutant cancer cells, which promotes the recruitment of M1 macrophages, neutrophils and myeloid-derived suppressor cells (40), likely predominates in the patient’s IMA microenvironment compared to the reference cohort, as demonstrated by the overexpression of myeloid lineage-related genes. Genes that regulate immune system processes and leukocyte activation, such as the dendritic cell marker CD1a and the chemokine CXCL13/BCA-1 were also overexpressed. CD1a is a marker for myeloid DC, which are a major source of IL-12 and Th1 CD4+ T cell polarization (41). Structurally related to the major histocompatibility complex (MHC) proteins, CD1a appears to be crucial for the presentation of non peptide tumour antigens to the immune system (42). Moreover, the density of CD1a+DCs within human tumors has been associated with survival (43).
CXCL13/BCA-1 has a controversial role in lung cancer. It may be induced by carcinogens and favor cancer cell proliferation (44), but it has also been proven to be fundamental to the development of tumor-associated lymphoid structures, which are detected in the patient’s tumor, and have been associated with a favorable prognosis in NSCLC (45).
Comparative transcriptomic analyses between the patient’s IMA and the IMA signature cases also revealed a limited range of genes with opposite expression pattern, mostly with an unclear function. Notably, HLA-DOA, CD74 and CIITA, encoding for MHCII molecules, which influence tumor antigen specific immune responses (46-48) were overexpressed in the patient’s IMA and downregulated in the IMA signature cases, whereas stemness and pro-tumoral genes, such as SOX4, HES1, IER3 and SERPINH1 (49-52) were downregulated in the patient’s IMA. This peculiar transcriptional profile is likely behind the favorable clinical course of the patient that, despite the 20.9 months of median survival for stage IV IMA (53).
The limits of this work lie in a transcriptional analysis that is confined to a single, albeit rare and unprecedented, clinical case. However, highlighting the value of both normal-like and cancer tissues in deciphering patient-specific tumor histogenesis, this study emphasizes the need for a personalized diagnostic and clinical management of the lung cancer patient.
Assessing the transcriptional profiles of both normal and neoplastic lung tissues provides fundamental information: (I) to define the case specific molecular pathways driving the natural history of a particular lung cancer histotype, and (II) to develop an accurate prognostic nomogram and patient-tailored treatment approach.
Beyond the long recognized clinical and prognostic value of the specific tumor histotype and stage, assessing the transcriptional signature of both normal and neoplastic lung tissues paves the way to personalized clinical treatment and outcome prediction.
Acknowledgments
Funding: The Project has been supported by two grants from the Italian Ministry of Health, Ricerca Finalizzata (RF-2013-02357552 and RF-2016-02362022), and one grant from the Italian Association for Cancer Research (AIRC) (IG 2019 – ID. 23264) to EDC.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-177). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). A written informed consent was obtained from the patient for the publication of this case report and for any images.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Braakhuis BJ, Tabor MP, Kummer JA, et al. A genetic explanation of Slaughter's concept of field cancerization: evidence and clinical implications. Cancer Res 2003;63:1727-30. [PubMed]
- Spira A, Beane JE, Shah V, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med 2007;13:361-6. [Crossref] [PubMed]
- Cao B, Feng L, Lu D, et al. Prognostic value of molecular events from negative surgical margin of non-small-cell lung cancer. Oncotarget 2016;8:53642-53. [Crossref] [PubMed]
- Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium, clinical implications of multicentric origin. Cancer 1953;6:963-8. [Crossref] [PubMed]
- Shim HS, Kenudson M, Zheng Z, et al. Unique Genetic and Survival Characteristics of Invasive Mucinous Adenocarcinoma of the Lung. J Thorac Oncol 2015;10:1156-62. [Crossref] [PubMed]
- Casali C, Rossi G, Marchioni A, et al. A single institution-based retrospective study of surgically treated bronchioloalveolar adenocarcinoma of the lung: clinicopathologic analysis, molecular features, and possible pitfalls in routine practice. J Thorac Oncol 2010;5:830-6. [Crossref] [PubMed]
- Yoshizawa A, Motoi N, Riely GJ, et al. Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage I cases. Mod Pathol 2011;24:653-64. [Crossref] [PubMed]
- Warth A, Muley T, Meister M, et al. The novel histologic International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification system of lung adenocarcinoma is a stage-independent predictor of survival. J Clin Oncol 2012;30:1438-46. [Crossref] [PubMed]
- Kadara H, Wistuba II. Field cancerization in non-small cell lung cancer: implications in disease pathogenesis. Proc Am Thorac Soc 2012;9:38-42. [Crossref] [PubMed]
- Guo M, Tomoshige K, Meister M, et al. Gene signature driving invasive mucinous adenocarcinoma of the lung. EMBO Mol Med 2017;9:462-81. [Crossref] [PubMed]
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1-13. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545-50. [Crossref] [PubMed]
- Rami-Porta R, Asamura H, Travis WD, et al. Lung cancer - major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J Clin 2017;67:138-55.
- Lim W, Ridge CA, Nicholson AG, et al. The 8(th) lung cancer TNM classification and clinical staging system: review of the changes and clinical implications. Quant Imaging Med Surg 2018;8:709-18. [Crossref] [PubMed]
- Lakshmanan I, Ponnusamy MP, Macha MA, et al. Mucins in lung cancer: diagnostic, prognostic, and therapeutic implications. J Thorac Oncol 2015;10:19-27. [Crossref] [PubMed]
- Cook DG, Strachan DP. Health effects of passive smoking-10: Summary of effects of parental smoking on the respiratory health of children and implications for research. Thorax 1999;54:357-66. [Crossref] [PubMed]
- Brody JS. Transcriptome alterations induced by cigarette smoke. Int J Cancer 2012;131:2754-62. [Crossref] [PubMed]
- Cao S, Yang C, Gan Y, et al. The Health Effects of Passive Smoking: An Overview of Systematic Reviews Based on Observational Epidemiological Evidence. PLoS One 2015;10:e0139907. [Crossref] [PubMed]
- Rager JE, Suh M, Chappell GA, et al. Review of transcriptomic responses to hexavalent chromium exposure in lung cells supports a role of epigenetic mediators in carcinogenesis. Toxicol Lett 2019;305:40-50. [Crossref] [PubMed]
- Agency for Toxic Substances and Disease Registry (ATSDR). Available online: https://www.atsdr.cdc.gov. Accessed October 11, 2019.
- Li J, Yang F, Li X, et al. Characteristics, survival, and risk factors of Chinese young lung cancer patients: the experience from two institutions. Oncotarget 2017;8:89236-44. [Crossref] [PubMed]
- Liu Q, Li A, Tian Y, et al. The CXCL8-CXCR1/2 pathways in cancer. Cytokine Growth Factor Rev 2016;31:61-71. [Crossref] [PubMed]
- Pine SR, Mechanic LE, Enewold L, et al. Increased levels of circulating interleukin 6, interleukin 8, C-reactive protein, and risk of lung cancer. J Natl Cancer Inst 2011;103:1112-22. [Crossref] [PubMed]
- Lagiou P, Trichopoulos D. Inflammatory biomarkers and risk of lung cancer. J Natl Cancer Inst 2011;103:1073-5. [Crossref] [PubMed]
- Luppi F, Longo AM, de Boer WI, et al. Interleukin-8 stimulates cell proliferation in non-small cell lung cancer through epidermal growth factor receptor transactivation. Lung Cancer 2007;56:25-33. [Crossref] [PubMed]
- Rot A. Contribution of Duffy antigen to chemokine function. Cytokine Growth Factor Rev 2005;16:687-94. [Crossref] [PubMed]
- Bonecchi R, Graham GJ. Atypical Chemokine Receptors and Their Roles in the Resolution of the Inflammatory Response. Front Immunol 2016;7:224. [Crossref] [PubMed]
- Yang M, Chen T, Han C, et al. Rab7b, a novel lysosome-associated small GTPase, is involved in monocytic differentiation of human acute promyelocytic leukemia cells. Biochem Biophys Res Commun 2004;318:792-9. [Crossref] [PubMed]
- Snider MD. A role for rab7 GTPase in growth factor-regulated cell nutrition and apoptosis. Mol Cell 2003;12:796-7. [Crossref] [PubMed]
- Li B, Huang Q, Wei GH. The Role of HOX Transcription Factors in Cancer Predisposition and Progression. Cancers (Basel) 2019;11:528. [Crossref] [PubMed]
- Li Q, Dong Z, Zhou F, et al. Krüppel-like factor 5 promotes lung tumorigenesis through upregulation of Sox4. Cell Physiol Biochem 2014;33:1-10. [Crossref] [PubMed]
- Tian J, Mahmood R, Hnasko R, et al. Loss of Nkx2.8 deregulates progenitor cells in the large airways and leads to dysplasia. Cancer Res 2006;66:10399-407. [Crossref] [PubMed]
- Kendall J, Liu Q, Bakleh A, et al. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci USA 2007;104:16663-8. [Crossref] [PubMed]
- Hsu DS, Acharya CR, Balakumaran BS, et al. Characterizing the developmental pathways TTF-1, NKX2-8, and PAX9 in lung cancer. Proc Natl Acad Sci USA 2009;106:5312-7. [Crossref] [PubMed]
- Duruisseaux M, Antoine M, Rabbe N, et al. Lepidic predominant adenocarcinoma and invasive mucinous adenocarcinoma of the lung exhibit specific mucin expression in relation with oncogenic drivers. Lung Cancer 2017;109:92-100. [Crossref] [PubMed]
- Guerra C, Mijimolle N, Dhawahir A, et al. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell 2003;4:111-20. [Crossref] [PubMed]
- Guerrero S, Casanova I, Farré L, et al. K-ras codon 12 mutation induces higher level of resistance to apoptosis and predisposition to anchorage-independent growth than codon 13 mutation or proto-oncogene overexpression. Cancer Res 2000;60:6750-6. [PubMed]
- Dias Carvalho P, Guimarães CF, Cardoso AP, et al. KRAS Oncogenic Signaling Extends beyond Cancer Cells to Orchestrate the Microenvironment. Cancer Res 2018;78:7-14. [Crossref] [PubMed]
- Cernadas M, Lu J, Watts G, Brenner MB. CD1a expression defines an interleukin-12 producing population of human dendritic cells. Clin Exp Immunol 2009;155:523-3. [Crossref] [PubMed]
- Brigl M, Brenner MB. CD1: antigen presentation and T cell function. Annu Rev Immunol 2004;22:817-90. [Crossref] [PubMed]
- Coventry B, Heinzel S. CD1a in human cancers: a new role for an old molecule. Trends Immunol 2004;25:242-8. [Crossref] [PubMed]
- Kazanietz MG., Durando M, Cooke M. CXCL13 and Its Receptor CXCR5 in Cancer: Inflammation, Immune Response, and Beyond. Front Endocrinol (Lausanne) 2019;10:471. [Crossref] [PubMed]
- Dieu-Nosjean MC, Antoine M, Danel C, et al. Long-term survival for patients with non-small-cell lung cancer with intratumoral lymphoid structures. J Clin Oncol 2008;26:4410-7. [Crossref] [PubMed]
- Thibodeau J, Bourgeois-Daigneault MC, Lapointe R. Targeting the MHC Class II antigen presentation pathway in cancer immunotherapy. Oncoimmunology 2012;1:908-16. [Crossref] [PubMed]
- Su H, Na N, Zhang X, Zhao Y. The biological function and significance of CD74 in immune diseases. Inflamm Res 2017;66:209-16. [Crossref] [PubMed]
- Accolla RS, Ramia E, Tedeschi A, et al. CIITA-Driven MHC Class II Expressing Tumor Cells as Antigen Presenting Cell Performers: Toward the Construction of an Optimal Anti-tumor Vaccine. Front Immunol 2019;10:1806. [Crossref] [PubMed]
- Vervoort SJ, van Boxtel R, Coffer PJ. The role of SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: friend or foe? Oncogene 2013;32:3397-409. [Crossref] [PubMed]
- Liu ZH, Dai XM, Du B. Hes1: a key role in stemness, metastasis and multidrug resistance. Cancer Biol Ther 2015;16:353-9. [Crossref] [PubMed]
- Arlt A, Schäfer H. Role of the immediate early response 3 (IER3) gene in cellular stress response, inflammation and tumorigenesis. Eur J Cell Biol 2011;90:545-52. [Crossref] [PubMed]
- Kamikawaji K, Seki N, Watanabe M, et al. Regulation of LOXL2 and SERPINH1 by antitumor microRNA-29a in lung cancer with idiopathic pulmonary fibrosis. J Hum Genet 2016;61:985-93. [Crossref] [PubMed]
- Cha YJ, Kim HR, Lee HJ, et al. Clinical course of stage IV invasive mucinous adenocarcinoma of the lung. Lung Cancer 2016;102:82-8. [Crossref] [PubMed]


