
Predicting the invasiveness of lung adenocarcinomas appearing as ground-glass nodule on CT scan using multi-task learning and deep radiomics
Introduction
Lung cancer is the most common cancer and the leading cause of cancer-related death (1). With the popularization of lung cancer screening with CT, the detection rate of pulmonary nodules is getting higher (2). Most of the early-stage lung cancers appear as ground glass opacity (GGN) on thin section CT. GGNs are classified into two categories according to the presence of solid components or not, pure ground-glass nodules (pGGNs) and mixed ground glass nodules (mGGNs) (3). According to the multidisciplinary classification of lung adenocarcinomas by the International Association for Lung Cancer Research, the American Thoracic Society and the European Society of Respiratory Sciences (IASLC/ATS/ERS) in 2011 (4), lung adenocarcinoma can be pathologically classified into four subtypes, atypical adenomatous hyperplasia (AAH), adenocarcinoma in situ (AIS), minimally invasive adenocarcinoma (MIA) and invasive adenocarcinoma (IAC). AAH and AIS is regarded as pre-invasive lesions.
Different subtypes of adenocarcinoma vary the clinical management strategies, survival rates, surgical approaches and postoperative therapeutic protocols. Pre-invasive lesions and MIAs often have good biological behavior, long-term unchanged or slow growth, and can be clinically followed up to select the best surgical timepoint, reducing the overtreatment (5,6). In contrast, IAC needs timely surgical treatment. The disease-free survival rate after surgery for patients with AAH/AIS/MIA can be close to 100%, which is significantly higher than that of IAC (38–86%, P<0.001), which depends mainly on different subtypes of IAC (7,8). Due to the high survival rate of MIA, pre-invasive lesions and MIA is defined as the ‘non-invasive’ adenocarcinoma. For the ‘non-invasive’ adenocarcinoma, sub-lobar resection is usually used to achieve the radical effect, and sub-lobar resection can retain more lung function, reduce postoperative complications and shorten recovery time (9). While for patients with IAC, lobectomy and mediastinal lymph node dissection are performed; moreover, if the postoperative adjuvant treatment is applied for those, the survival rate may be improved, which is of great significance for the individualization of treatment (10,11). Therefore, evaluation of invasiveness of lung nodules is important to select the appropriate clinical-decision strategy.
Although intraoperative freezing plays a great role for the evaluation of the invasiveness of lung nodules, it is limited due to a variety of reasons, such as uneven diagnostic levels, clinical experience of pathologists, frozen materials and technical conditions during surgery, inaccurate localization of the lesions, inaccurate material selection, too small lesions, and the complications (12,13). Therefore, it is more necessary to judge the invasiveness of GGNs before surgery, CT as a non-invasive method plays a great role in the preoperative evaluation the invasiveness of GGNs. Unfortunately, it has been a great challenge to distinguish the different histological invasiveness with traditional CT morphological findings. Currently, morphological findings are most commonly used to differentiate invasive from non-invasive adenocarcinoma in clinical work, such as proportion of solid component volume, non-smooth margin, lobulation and nodule size. Considerable overlapping in CT morphological features among various histological subtypes have been reported (14,15). It is difficult to differentiate histological invasiveness with the morphological features alone.
With the arrival of the era of big data, many studies have focused on radiomics and pathological subtypes of lung adenocarcinomas. Although the accuracy of radiomics is higher than that of traditional morphological signs (16), the conventional radiomic methods are tedious and time-consuming. In recent years, artificial intelligence (AI) is an emerging concept in computer science research. Deep neural networks (DNN) has achieved broad application in many domains, especially in the analysis of medical images, including images of skin lesions, pneumonia, and clinical pathological images (17,18). Deep learning has better performance in many clinical aspects than traditional qualitative analysis tools. As yet, few studies assess the performance of a model combining a deep convolutional neural network and a hand-crafted radiomics signature to differentiate the histological invasiveness of lung adenocarcinomas manifesting as GGNs. The study aims to build three deep learning models, then compare the performance of the models on the invasiveness classification of GGNs based on chest CT. Besides, the performance of our models was compared with quantitative analysis of maximum nodule diameters. The authors have completed the STROBE reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-370).
Methods
Study population
From January 2012 to March 2018, 794 patients with lung adenocarcinoma showing GGNs were enrolled in this retrospective study. The included patients were those with (I) no previous therapy history before CT examination; (II) lung adenocarcinoma confirmed by surgical resection and histopathological diagnosis; (III) tumor less than 3cm in diameter on thin-slice (0.625–1 mm) CT images. The exclusion criteria were as follows: (I) marked artifacts on CT images; (II) history of preoperative treatment; (III) incomplete clinical information or DICOM images; (IV) history of other malignant tumors; (V) lung cancer associated with cystic airspaces. The patients were divided into three categories of pre-invasive lesions, MIA and IAC according to pathological examination results. Moreover, “non-invasive” adenocarcinoma and IAC were also made a binary classification. Three different classification tasks were considered: (I) classification of AAH/AIS with MIA; (II) classification of MIA with IAC; (III) classification of AAH/AIS&MIA with IAC. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the Ethics Committee of the Changzheng Hospital, Second Military Medical University (No. 2018SL049). Because of the retrospective nature of the research, the requirement for informed consent was waived.
All patients underwent non-enhanced CT scanning with one of the five scanners in our hospital, as described in our previous study (19). All patients took the supine position and adopted the whole lung scan at the end of inspiration. Multi-planner reconstruction (MPR) was used for images reconstruction with thin-slice (≤1 mm) images. The demographic data including age and gender were derived from medical records. All patients were diagnosed with the same manner of histopathological diagnosis after surgical resection. The pathological subtypes of each GGNs were categorized according to the IASLC/ATS/ERS classification of lung adenocarcinoma in 2011 (4).
Dataset preprocess
GGNs were divided with the ratio of 0.7:0.15:0.15 for all training dataset, validation dataset and the test dataset (Table 1). However, the number of IAC patients exceed the sum number of AAHs and AISs. This imbalanced data distribution may have a direct effect on the performance of the deep neural networks. In order to alleviate such effect, AAH and AIS were regarded as the one category.
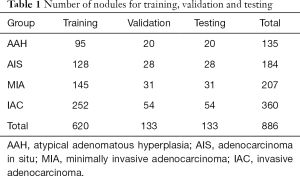
Full table
Nodule labeling and segmentation
The segmentation of volume of interest (VOI) of each nodule was delineated manually and independently by experienced thoracic radiologist with lung window settings (window width 1,500 HU, window level 450 HU) using self-developed software from Shanghai Aitrox Information Technology Co., Ltd. The GGNs largest transverse cross-sectional diameter was measured in the lung window as the maximum nodule diameter. Large vessels and bronchi were excluded manually from each VOI. VOIs were marked with specific labels (AAH, AIS, MIA, and IAC) according to the pathological reports.
Building three deep-learning models
3D Image Patch generation and radiomic feature extraction
The two end-to-end networks, XimaNet and XimaSharp, directly take 3D image patches as input. To generate the 3D image patches, we firstly resampled the image spacing to 1 mm per pixel in all three dimensions, and normalized the CT value in each pixel to [−1, 1]. To keep the redundant image information around the tumor lesion, which could be meaningful for lung nodule invasiveness classification, we did not directly crop the lesion according to its corresponding mask. Instead, we calculated the maximal diameter of the lesion in all three dimensions. We cropped a 3D image patch around the lesion with twice the maximal diameter in all three dimensions and resized it to 64×64×64 pixels. Especially, if the maximal diameter is smaller than 32 pixels, we directly cropped a 64×64×64 3D image patch around the lesion without resizing.
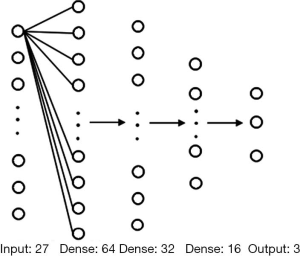
In contrast, the non-end-to-end network, deep-RadNet, takes selected radiomic features instead of image patches as input. We used the radiomic extracting tool PyRadiomics (20) (https://pyradiomics.readthedocs.io/en/latest/) to extract the radiomic features from the Region of Interest (ROI) on the images, which was labelled by corresponding masks. From the 1,743 extracted radiomic features, we selected the ones most relevant to the lung nodule invasiveness by: (I) excluding 5% abnormal samples with isolation forest (IF) algorithm (21); (II) deleting the features with low variance; (III) calculating the z-scores for each feature among all samples (mean normalization); (IV) reducing the feature numbers by automatic relevance determination (ARD) (22) and least absolute shrinkage and selection operator (LASSO) (23) with 10-fold cross validation. Finally, a number of 27 radiomic features were selected during the selection process.
Buliding the XimaNet and XimaSharp architecture
The XimaNet design was inspired by the ResNet (24) structure. The network structure was shown in Figure 1A. It took 64×64×64 pixel image patches as input, and the image patches were batch normalized, went through 64 convolutional layers and batch normalized again. Then the patches went through 6 building block modules, whose structure was shown in Figure 1B. The stride of the first building block was 1, and the rest building blocks had stride =2 for down-sampling. The building blocks exported a 2×2×2 pixel feature map, which was passed through a module consisting of batch normalization (BN) (25) and rectified linear unit (ReLU) (26). Then the feature map went through global average pooling (GAP) and dense layers to output three predicted probability values corresponding to AAH/AIS, MIA and IAC respectively. The final prediction was the category with the maximal predicted probability. During the training process, we applied data augmentation by flipping the images on x-axis only, y-axis only and both axes simultaneously. This DL network was trained based on TensorFlow 1.10.0 (27) and Keras 2.2.4 with Python 2.7, on a workstation equipped with 2 NVIDIA 1080Ti GPUs.
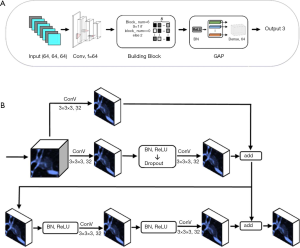
The XimaSharp network was inspired by DenseSharp network (28). It was a multitask network for simultaneous classification and segmentation. The basic structure of XimaSharp was similar to XimaNet, but we up-sampled the 2×2×2 pixel feature map exported by the building blocks to feature maps of 4×4×4, 8×8×8, 16×16×16, 32×32×32 and 64×64×64 pixels and added them to the feature maps exported by each building block of the corresponding size. In the end, we exported a segmentation mask with the size of 64×64×64 pixels and evaluated the model performance by calculating both the classification and segmentation loss. The XimaSharp model was trained under the same environment to XimaNet.
Building deep-RadNet architecture
The deep-RadNet took the 27 selected radiomic features as input. We used three fully connected layers before the model exported the final prediction. Similar to the output of XimaNet, the prediction was also three probability values corresponding to AAH/AIS, MIA, and IAC, and the category with the maximal probability was the final prediction. The structure of deep-RadNet was shown in Figure 2.
Training of models for classification of GGNs
We applied the cross-entropy function as the loss function for XimaNet and deep-RadNet. The formula is shown below:
[1]
In which tcls is the ground truth label, ycls is the prediction result from our model, n is the sample number, and c is a class. For our classification and segmentation multitask model XimaSharp, the loss function consists of both classification loss and segmentation loss. The formula is shown below:
ljoint = lcls + λlseg[2]
The parameter λ indicates the weight for segmentation loss. In our case, it is set as 0.2 as an empirical value. lsegis the dice loss for segmentation whose formula is shown below:
[3]
In which tseg is the manually labelled ground truth mask and yseg is the mask predicted by the model, and n is the total number of the samples. In the training process, we used Adam optimizer (29). The learning rate was set to 0.01, and the decay rate was set to 0.334. The dropout was set to 0.5 in the first building block, and 0.3 in all the other blocks. The models were trained for 80 epochs. For the deep-RadNet, we used the same cross entropy loss function as the one for XimaNet. The optimizer was Adam, and the learning rate was set to 2×10−4. The decay was set to 1×10−6. The model was trained for 80 epochs.
Evaluation of model performance
The input for XimaNet and XimaSharp was a 64×64×64 pixel image patch. XimaNet was used to predict the classification of AAH/AIS, MIA and IAC, while XimaSharp was used to predict the invasiveness degree as well as lesion segmentation mask. F1-score was used to assess the accuracy of the three-category classification mode. The maximum value of F1 score was 1 and the minimum value was 0. The “weighted average F1-score” was performed to reduce the effect of imbalanced data. The “weighted average F1-score” is a transformation of F1-score which is calculated from various kinds of F1 weighted calculation. The formulas are shown below:
[4]
[5]
We also used the Matthews correlation coefficient (MCC) for model evaluation, which was insensitive to unbalanced data. The formula for MCC is shown below:
[6]
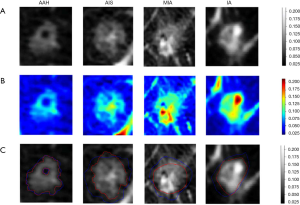
To understand the “black boxes” of the deep learning model, heat maps were generated by Grad-CAM to visualize the most indicative region for the invasiveness of GGNs. Grad-CAM was a gradient related distribution map which could visualize the significance of the region the algorithm focused on.
Statistical analysis
All statistical analysis were performed in Matlab (version 2019a; MathWorks, Narick, Mass). Receiver operating characteristic curves (ROCs) as well as areas under receiver operating characteristic curves (AUCs) were used to assess overall classification performance of the three models. Then, Z test was applied to evaluate the difference of performance among models. Bootstrapping (1,000 boot-strap samples) was used to calculate 95% CIs and the associated P values. P<0.05 was considered a statistically significant difference. The performance by the size of GGNs was evaluated by t test. The optimal cut-off diameter for GNNs classification was calculated by searching in the dataset to maximize accuracy. Double tail distribution and double sample equal variance hypothesis were selected for parameters for tails and type, and P values were calculated under the optimal cut-off size.
Results
A total of 794 patients with 886 lung nodules were evaluated. The patient characteristics were illustrated in Supplementary file. We evaluated the performance in three classification tasks of the three DNNs: XimaNet, XimaSharp and deep-RadNet. Evaluation metrics included accuracy, “weighted average F1-score” and Matthews correlation coefficient (MCC), the classification performance was shown in Table 2. Table 2 showed deep-RadNet presented with the highest accuracy, “weighted average F1-score” and MCC in comparison with other two models in all the three classification tasks. The AUC for Deep-RadNet was 0.891, 0.889, 0.941for the AAH/AIS and MIA classification, the MIA and IAC classification, and AAH/AIS/MIA and IAC classification, respectively, which were higher than those of other two models (Figure 3).
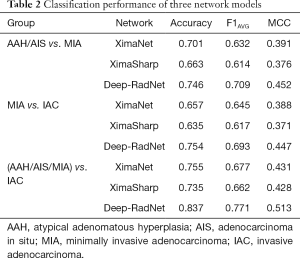
Full table

The results showed that nodule size was a significant differentiator of non-invasive nodules from invasive nodules (P=0.04, accuracy =0.701, include AAH, 1 cm; P=0.01, accuracy =0.72, exclude AAH, 2 cm). The accuracy indicated that the classification ability of deep_RadNet was beyond that of the lesion size to differentiate histological invasiveness by the optimal cut-off diameter.
Moreover, the Z-test was used to compare the performance among 3 models. P values of the Z-test was 0.021(P<0.05) between deep_RadNet and XimaNet, 0.019 (P<0.05) between deep_RadNet and XimaSharp and 0.98 (P>0.05) between XimaNet and XimaSharp, which indicated that the deep_RadNet revealed the best performance.
Discussion
The invasiveness of GGNs is associated with disease prognosis, choice of therapeutic approach, and reduction of overtreatment. This study showed deep learning system combined with the radiomics features could conveniently and automatically obtain the best performance in predicting the invasiveness of lung adenocarcinoma manifesting as GGNs, in comparison with other two models. To the best of our knowledge, this study was the first to present a XimaSharp model to detect and segment GGNs automatically, and a Deep-RadNet model to evaluate the invasiveness of GGNs accurately.
There are reports claiming that the optimal cut-off diameter is helpful for evaluating the degree of invasiveness of lung adenocarcinoma. There is no consensus among different studies in that distinguishing the invasiveness degree referring to the sizes of GGNs merely. Lee et al. reported that 14 mm was the optimal cut-off value to differentiate pre/minimally invasive from IAC, and the sensitivity of 67% and a specificity of 74% (14). Lim et al. suggested that the total tumor size of 10 mm could act as a direct criterion to distinguish pre-invasive and IAC (30). Our study showed that the AUC values of lesion size in differentiating non-invasive nodules form invasive nodules were 70.1% with the cutoff value of 10 mm. The optimal cut-off value was 2cm for differentiating non-invasive adenocarcinoma from IAC when the AAH was excluded. The differences can be explained by two reasons. The first reason is that the AAH has not been included in some research. The second is that the definition of nodule size is not uniform. Therefore, the doctor judged the invasiveness of pulmonary nodules by the optimal cut-off value remains controversial.
It has been reported that the solid components in the GGN plays a great role in the differentiation of invasiveness. The solid components on CT cannot evaluated accurately, due to the different pathological features with the similar solid component on CT images. Solid component may be the proliferation of fibroblasts and/or the invasive components of tumor cells, indicating a benign scar or collapse of the alveolar wall or tumor (31). Besides, there are lots of overlapping in the morphological characteristics of non-invasive nodules and invasive ones. Therefore, it is limited to differentiate the invasiveness degree by some morphological characteristic of GGNs.
For the traditional deep learning method, it was difficult to extract features in the training process of the classification model. In contrast, traditional radiomics method extracts interpretative features from medical images based on prior medical knowledge combined with image processing methods. After feature extraction, features that contributed most to the prediction were selected and fed into the traditional statistical model for classification results. The critical idea of our Deep-RadNet model was to choose features that contributed most to the prediction results and fed them into deep learning neural network for training, which may improve the prediction accuracy and lead to better clinical interpretability comparing to a traditional radiomics model. We explored the possibility to combine radiomics and deep learning network for lung nodule invasiveness classification tasks, and showed the best performance.
We also attempted to explore a deep learning model for lung nodule segmentation tasks, just as shown in Figure 4. The heatmap demonstrated that the most meaningful region for differentiating the degree of invasiveness was the solid component inside the tumor. This finding highlighted the importance of this region, which may indicate the most invasive tumor components and assist radiologists in making accurate judgments of invasiveness of GGNs.
This study has some limitations. First is the limited data accessibility due to a single-center study, which may lead to more selection bias and compromise in the generalization ability of our classifier. However, five independent scanners in our hospital were used and all the three models were found to be reproducible in the validation and training groups. Second, our models depended on either the pre-defined radiomic features or image features automatically extracted by deep learning algorithm for the classification, while some traditional morphological characteristics such as spiculation and lobulation were not considered. In the future studies, such information should be combined with radiomic features for building a more accurate lung-nodule invasiveness prediction model. Also, considering the recent progress in lung nodule diagnosis with contrast enhanced CT (32), we could extend to contrast enhanced CT images and investigate its performance for the invasiveness classification of GGNs.
Conclusions
In conclusion, the deep learning model integrating the radiomics features of GGNs with the visual heat map could evaluate the invasiveness of GGNs accurately and intuitively, which is convenient for clinicians, showing great potential to improve the efficiency of lung cancer screening and providing a theoretical basis for individualized and accurate medical treatment of patients with GGNs.
Acknowledgments
Funding: The National Key Research and Development Program of China for Intergovernmental Cooperation (grant number 2016YFE0103000); Shanghai Municipal Commission of Health and Family Planning Program (grant number 2018ZHYL0101); Technology Commission of Shanghai Municipality (grant number 17411952400); The key project of the National Natural Science Foundation of China (grant number 81930049); National Key Research and Development Project (grant number 2018YFC0116404); National Natural Science Foundation of China (grant number 81871321), National Key R&D Program of China (grant number 2017YFC1308703).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-370
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-370
Peer Review File: Available at http://dx.doi.org/10.21037/tlcr-20-370
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-370). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the Ethics Committee of the Changzheng hospital, Second Military Medical University (No.2018SL049). Because of the retrospective nature of the research, the requirement for informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Goo JM, Park CM, Lee HJ. Ground-glass nodules on chest CT as imaging biomarkers in the management of lung adenocarcinoma. AJR Am J Roentgenol 2011;196:533-43. [Crossref] [PubMed]
- Yang J, Wang H, Geng C, et al. Advances in intelligent diagnosis methods for pulmonary ground-glass opacity nodules. Biomed Eng Online 2018;17:20. [Crossref] [PubMed]
- Travis WD, Brambilla E, Noguchi M, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society International Multidisciplinary Classification of Lung Adenocarcinoma. J Thorac Oncol 2011;6:244-85. [Crossref] [PubMed]
- Jia M, Yu S, Cao L, et al. Clinicopathologic Features and Genetic Alterations in Adenocarcinoma In Situ and Minimally Invasive Adenocarcinoma of the Lung: Long-Term Follow-Up Study of 121 Asian Patients. Ann Surg Oncol 2020;27:3052-63. [Crossref] [PubMed]
- Soda H, Nakamura Y, Nakatomi K, et al. Stepwise progression from ground-glass opacity towards invasive adenocarcinoma: long-term follow-up of radiological findings. Lung Cancer 2008;60:298-301. [Crossref] [PubMed]
- Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. [Crossref] [PubMed]
- Yanagawa N, Shiono S, Abiko M, et al. New IASLC/ATS/ERS classification and invasive tumor size are predictive of disease recurrence in stage I lung adenocarcinoma. J Thorac Oncol 2013;8:612-8. [Crossref] [PubMed]
- Ye T, Deng L, Wang S, et al. Lung Adenocarcinomas Manifesting as Radiological Part-Solid Nodules Define a Special Clinical Subtype. J Thorac Oncol 2019;14:617-27. [Crossref] [PubMed]
- Russell PA, Wainer Z, Wright GM, et al. Does lung adenocarcinoma subtype predict patient survival?: A clinicopathologic study based on the new International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary lung adenocarcinoma classification. J Thorac Oncol 2011;6:1496-504. [Crossref] [PubMed]
- Ettinger DS, Akerley W, Borghaei H, et al. Non-small cell lung cancer, version 2.2013. J Natl Compr Canc Netw 2013;11:645-53. [Crossref] [PubMed]
- Annema JT, Veselic M, Versteegh MI, et al. Mediastinal restaging: EUS-FNA offers a new perspective. Lung Cancer 2003;42:311-8. [Crossref] [PubMed]
- Huang MD, Weng HH, Hsu SL, et al. Accuracy and complications of CT-guided pulmonary core biopsy in small nodules: a single-center experience. Cancer Imaging 2019;19:51. [Crossref] [PubMed]
- Lee SM, Park CM, Goo JM, et al. Invasive pulmonary adenocarcinomas versus preinvasive lesions appearing as ground-glass nodules: differentiation by using CT features. Radiology 2013;268:265-73. [Crossref] [PubMed]
- Sakurai H, Nakagawa K, Watanabe S, et al. Clinicopathologic features of resected subcentimeter lung cancer. Ann Thorac Surg 2015;99:1731-8. [Crossref] [PubMed]
- Fan L, Fang M, Li Z, et al. Radiomics signature: a biomarker for the preoperative discrimination of lung invasive adenocarcinoma manifesting as a ground-glass nodule. Eur Radiol 2019;29:889-97. [Crossref] [PubMed]
- Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115-8. [Crossref] [PubMed]
- Ragab DA, Sharkas M, Marshall S, et al. Breast cancer detection using deep convolutional neural networks and support vector machines. PeerJ 2019;7:e6201. [Crossref] [PubMed]
- Wang X, Zhao X, Li Q, et al. Can peritumoral radiomics increase the efficiency of the prediction for lymph node metastasis in clinical stage T1 lung adenocarcinoma on CT? Eur Radiol 2019;29:6049-58. [Crossref] [PubMed]
- van Griethuysen JJ, Fedorov A, Parmar C, et al. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res 2017;77:e104-7. [Crossref] [PubMed]
- Liu FT, Kai MT, Zhou ZH, editors. Isolation Forest. Available online: https://cs.nju.edu.cn/zhouzh/zhouzh.files/publication/icdm08b.pdf?q=isolation-forest
- Wipf DP, Nagarajan SS, editors. A New View of Automatic Relevance Determination. Advances in Neural Information Processing Systems 20, Proceedings of the Twenty-First Annual Conference on Neural Information Processing Systems. British Columbia, Canada: Vancouver, 2007.
- Tibshirani R. Regression Shrinkage and Selection Via the Lasso. J R Stat Soc 1996;58:267-88.
- He K, Zhang X, Ren S, et al. (eds). Deep Residual Learning for Image Recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2016.
- Ioffe S, Szegedy C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. Available online: http://proceedings.mlr.press/v37/ioffe15.pdf
- Hara K, Saito D, Shouno H. Analysis of function of rectified linear unit used in deep learning. 2015 International Joint Conference on Neural Networks (IJCNN); 12-17 July 2015; Killarney, Ireland. IEEE, 2015. doi: 10.1109/IJCNN.2015.7280578. [Crossref]
- Abadi M, Barham P, Chen J, et al. TensorFlow: A system for large-scale machine learning. Available online: https://www.usenix.org/system/files/conference/osdi16/osdi16-abadi.pdf
- Yang J, Fang R, Ni B, et al. Probabilistic Radiomics: Ambiguous Diagnosis with Controllable Shape Analysis. Medical Image Computing and Computer Assisted Intervention (2019). doi: 10.1007/978-3-030-32226-7_73. [Crossref]
- Kingma D, Ba J. Adam: A Method for Stochastic Optimization. Available online: https://arxiv.org/pdf/1412.6980.pdf
- Lim HJ, Ahn S, Lee KS, et al. Persistent pure ground-glass opacity lung nodules >/= 10 mm in diameter at CT scan: histopathologic comparisons and prognostic implications. Chest 2013;144:1291-9. [Crossref] [PubMed]
- Lee KH, Goo JM, Park SJ, et al. Correlation between the size of the solid component on thin-section CT and the invasive component on pathology in small lung adenocarcinomas manifesting as ground-glass nodules. J Thorac Oncol 2014;9:74-82. [Crossref] [PubMed]
- Liu Y, Liu S, Qu F, et al. Tumor heterogeneity assessed by texture analysis on contrast-enhanced CT in lung adenocarcinoma: association with pathologic grade. Oncotarget 2017;8:53664-74. [Crossref] [PubMed]


