
Longitudinal multi-gene panel assessment of circulating tumor DNA revealed tumor burden and molecular characteristics along treatment course of non-small cell lung cancer
Introduction
Lung cancer is the leading cause of cancer death worldwide (1). For patients with advanced stage lung cancer, treatment options include conventional systemic chemotherapy or molecular-targeted therapy, and the latter is indicated with identification of therapeutic tumor targets (2). Actionable tumor targets, such as epidermal growth factor receptor (EGFR) mutations and anaplastic lymphoma kinase (ALK) translocation, are present more commonly in lung adenocarcinomas in Asians, and the presence of these actionable targets indicate that the tumour would be sensitive to specific tyrosine kinase inhibitors (TKIs) (2). Therefore, the identification of actionable mutations in tumors can guide treatment choice. Nevertheless, it is often impractical to obtain tumor tissue biopsy for molecular profiling, let alone to repeat tissue samplings over time to evaluate acquired resistance due to further genetic aberrations. The liquid biopsy approach, which offers the option of detecting molecular characteristics of circulating tumor cells or circulating tumor DNA (ctDNA) in blood (3), has the potential to revolutionize clinical care in cancer patients (2). Longitudinal follow up with liquid biopsy may be a new way to inform clinicians of subtle changes in underlying tumor characteristics or early knowledge of disease progression. This may allow for discovery of acquisition of oncogenic mutations that could be biomarkers for the next line of anti-cancer treatment or for prognostication (4,5).
ctDNA is cell-free DNA released from tumor cells into the circulation (6). Our group had previously shown the feasibility of detecting concordant EGFR mutations in plasma and tumor tissue of advanced stage lung cancer patients, and the presence or increasing plasma levels of ctDNA was associated with a worse prognosis (7). Similarly, it has been reported in other studies that somatic mutations in ctDNA of patients with non-small cell lung cancer (NSCLC) reflect molecular characteristics in tumor tissue, and ctDNA levels correlated with tumor stage and tumor burden (8-10). These observations have prompted subsequent studies to examine if ctDNA testing is useful for monitoring tumor status and tumor response to therapy over time. However, most of these previous studies tested for either a single gene (e.g., EGFR) or a few genes in ctDNA (11-13) or when a larger panel of genes was used, ctDNA was only examined at one single time point (14,15). In this proof-of-concept study, we employed a longitudinal design and a panel of 64 genes to examine whether (I) serial testing of ctDNA over time in the same patient was feasible for monitoring tumor burden, and (II) sequencing a panel of genes, as compared to the single gene approach, provided additional advantage in revealing tumor status and acquisition of new ctDNA mutations along treatment course. We present the following article in accordance with the AME publishing reporting checklist.
The authors present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-675).
Methods
Study population
Consecutive patients with advanced stage lung cancer attending the clinics of the Department of Medicine, Queen Mary Hospital, were recruited. The inclusion criteria were: (I) patients had diagnosis of advanced stage NSCLC and would undergo anti-cancer treatment, with either first- or second-line EGFR-TKI for patients with EGFR mutations, or first line chemotherapy for patients with EGFR wildtype tumors; and (II) patients gave informed written consents and agreed to have study follow up with plasma samples taken at three time points along their course of treatment: enrollment/baseline, at stable disease, and upon clinical progression of disease according to the RECIST 1.1 criteria. Blood samples of at least 30 mL were collected at each clinical visit time-point along the course of treatment. Plasma and serum samples were processed within 1 hour of collection and stored at –80 °C. Clinical information of recruited subjects was recorded. At the time of enrollment, each participant also completed a questionnaire pertaining to their demographic information, lifestyle factors, and medical history. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study protocol was approved by the Ethics Committee of the University of Hong Kong and Hong Kong Hospital Authority Hong Kong West Cluster Institutional Review Board (IRB Reference Number UW 16-104). Informed consent was obtained from all the participants.
The first patient was recruited in April, 2016 and the last patient who completed all the follow-up with disease progression had the last blood sample collected in October 2018. Stable disease was confirmed with review of all the records for the 20 recruited subjects, with neither recent sufficient shrinkage to qualify for partial or complete response nor sufficient increase to qualify for progressive disease.
ctDNA measurements
For each patient, a plasma sample of at least 1 (median: 2, range: 1 to 4) mL, processed from EDTA tubes from each time point was shipped frozen overnight to Personal Genome Diagnostics (PGDx, Baltimore, MD, USA) for ctDNA analysis (16,17). Samples were labeled with a unique sample number, and serial samples of the same participant could not be identified by laboratory personnel. Circulating cell-free DNA was extracted and analyzed for a panel of 64 well-characterized cancer genes (PlasmaSELECTTM 64) by next generation sequencing with about 30,000× coverage (16,17). PlasmaSELECTTM 64 allows the identification of single nucleotide variants (SNVs), indels, amplifications, translocations, and microsatellite instability with sensitivity of 99.4% for SNVs and indels and a per base specificity of >99.9% (16,17). Somatic mutations in ctDNA (point mutations, insertion, deletion and substitution) were identified by the proprietary VariantDx bioinformatics pipeline that incorporates information from public databases, such as dbSNP, the 1000 Genome Project, and COSMIC, and excludes potential germline as well as hematopoietic variants (18-21). The total level of ctDNA in a sample was assessed as the total number of mutant fragments per mL of plasma. The ctDNA level of a particular gene (e.g., EGFR) was reported as the total number of mutant fragments detected in that gene per mL of plasma.
Statistical analysis
Our goal was to determine if mutation profiles in ctDNA reflected tumor burden over time and hence varied with tumor status at enrollment/baseline, stable disease, and progression. Statistical analysis focused on within-individual pairwise comparison of ctDNA profiles at any two of the three time points. Wilcoxon signed-rank test and exact binomial McNemar’s test were used for comparison of continuous variable (ctDNA quantitative level) and categorical variable (ctDNA detection), respectively. Reported P values are two-sided.
Results
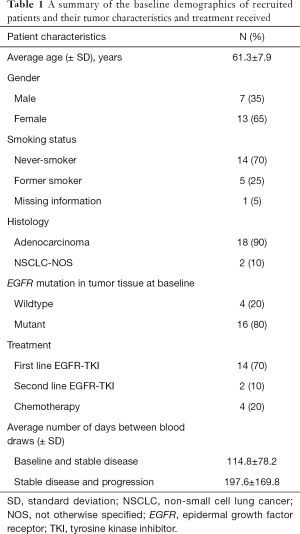
Twenty patients who fulfilled inclusion criteria were recruited and their data analyzed. Their demographics, disease staging, driver mutations, treatment, tumor measurements by the longest diameter, sites of metastasis and concentration of ctDNA at baseline, stable disease and progressive disease were all listed in Table 1. A summary of these patients’ baseline characteristics are shown in Table 2. All patients were of Chinese descent and had stage IV NSCLC. The majority of patients were females, never-smokers, diagnosed with adenocarcinoma of lung, and 16 of them (16/20, 80%) were tested positive for EGFR mutations in tumor tissue. The median time interval between ctDNA samples were 84 days [inter-quartile range (IQR): 53–193] between baseline and stable disease, 145 days (IQR: 80–252) between stable disease and progression, and 259 days (IQR: 171–376) between baseline and progression. Median cell-free DNA yield per mL of plasma sample was 20.5 ng (IQR: 12.4–26.2), and the median total yield was 37.3 ng (IQR: 28.7–51).
Full table
Full table
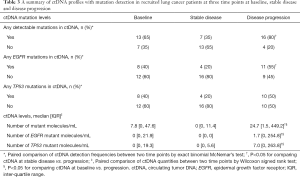
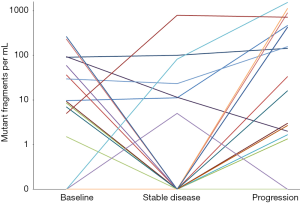
The detection frequencies of ctDNA in these 20 patients at the three time points are summarized in Table 3. Even in this cohort of patients with a high prevalence of EGFR driver mutations, more than 30% of patients tested positive for ctDNA have alterations in non-EGFR genes (38.5%, 42.9%, and 31.3% at baseline, stable disease, and progression, respectively). None of the patients had microsatellite instability, amplification, or rearrangement. In general, the proportion of patients with detectable ctDNA decreased from baseline (65%) to stable disease (35%), and rose to the highest at the time of progression (80%) (P=0.012, pairwise comparison of ctDNA detection between stable disease and progression). Similar patterns were observed for the detection of somatic EGFR and TP53 alterations over the three time points. ctDNA quantities in terms of the number of mutant fragments per mL of plasma are shown in Table 3. Median levels of ctDNA were 7.8 at baseline, zero at time of stable disease, and 24.7 at disease progression (pairwise difference: P=0.013 between baseline and progression; P=0.007 between stable disease and progression). ctDNA quantitative levels in individual patients over three time points are plotted in Figure 1.
Full table
The largest difference in ctDNA detection frequency was observed between stable disease and progression, within-subject results are detailed in Table 4. Using a 64-gene panel, we found that in 50% of the patients, ctDNA detection changed from negative at the time of stable disease to positive during disease progression. Contrarily, change in EGFR detection from absence to presence occurred in 35% of the patients when disease progressed (mostly in EGFR T790M and/or E746_A750del). Therefore, if a single gene of EGFR were assessed instead of using a multi-gene panel, the change in ctDNA pattern indicative of disease progression would have been missed in 15% of the patients who had ctDNA mutations in non-EGFR genes.

Full table
The detection of additional ctDNA mutations or their disappearance during treatment course in the 20 patients is detailed in Table 5. Based on their tumors being EGFR mutants or wildtype, and the serial changes in ctDNA mutation patterns, recruited patients could be broadly divided into four groups:
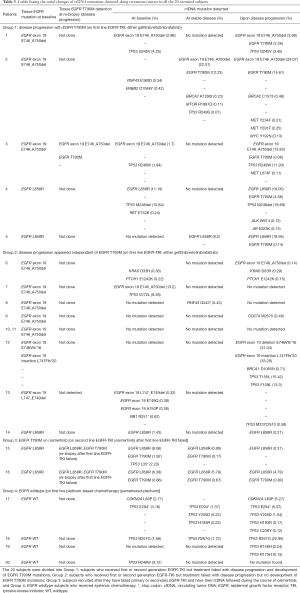
Full table
❖ Group 1 (patients 1–5): patients had an EGFR mutation, but not EGFR T790M, in tumor tissue at diagnosis, were treated with first line EGFR-TKI (gefitinib, erlotinib or afatinib) at baseline and had mainly EGFR T790M mutation detected at disease progression;
❖ Group 2 (patients 6–14): these patients were similar to Group 1 in terms of having had a drug-sensitive EGFR mutation in tumor tissue and were treated with EGFR-TKI, but they never had EGFR T790M mutation detected along their treatment course;
❖ Group 3 (patients 15 and 16): patients had sensitizing EGFR mutations and disease progression after first line EGFR-TKI, switched to second line EGFR-TKI (osimertinib) with baseline blood taken when they started osimertinib, and followed with stable disease then further disease progression on osimertinib;
❖ Group 4 (patients 17–20): EGFR wildtype subjects on first line platinum-based chemotherapy (pemetrexed-platinum) at baseline.
Among the 14 EGFR mutant subjects on first line EGFR-TKI treatment, 5 (5/14, 35.7%) (Group 1) had detection of EGFR T790M mutation at disease progression (Table 5). In these five subjects, other oncogenic mutations were also found at the time of disease progression, such as MYC and MET mutations. Nine of the 14 patients (64.3%) progressed without EGFR T790M mutation detected, but new ctDNA mutations were also found in various genes such as BRCA1, CD274 and TP53.
Two EGFR mutant patients (Group 3) showed persistent detection of EGFR T790M at osimertinib baseline through stable disease on treatment. At further disease progression on osimertinib, one of them (patient 15) lost the EGFR T790M in ctDNA while the other one (patient 16) showed persistent EGFR T790M mutation detection at further disease progression. Neither patient had new mutations detected at time of progression.
In some patients with either EGFR mutations or wildtype EGFR in tumor tissue, different mutations in the TP53 gene were detectable in ctDNA at different time-points. Nevertheless, TP53 mutations appeared to be more prevalent and were more likely to accumulate diverse types of point mutation in the wildtype EGFR tumors when the respective patients were treated with chemotherapy (Group 4).
Discussion
In this proof-of-concept longitudinal study using a multi-gene panel, we showed that ctDNA detection and quantity reflected tumor status over time—after initiation of treatment, levels decreased during stable disease and then increased again when disease progressed. Multi-gene ctDNA assessment also revealed mutations acquired and accumulated during anti-cancer treatment and reflected the underlying heterogeneous tumor biological characteristics.
Previous cross-sectional studies had found ctDNA levels to be positively associated with tumor stage, tumor burden, or tumor volume of lung cancer (8,10,22). In a study using CAPP-Seq for 139 genes, ctDNA detection increased from 50% in stage I NSCLC tumors to 100% among stage II–IV patients, and ctDNA levels correlated with tumor volume as measured by imaging (R2=0.89) (8). Because of the cross-sectional nature of these studies, they were not able to evaluate whether ctDNA changed with the evolution of tumor characteristics over time or with course of treatment. On the other hand, longitudinal studies on the utility of ctDNA in monitoring tumor status were mostly done with testing of EGFR gene only. In general, these studies showed that increased levels of plasma-mutant EGFR and detection of specific EGFR mutations in ctDNA at baseline were associated with poorer response to EGFR-TKI and poor progression-free and overall survivals (9,12,23). In serial samples, the levels of ctDNA containing EGFR mutations usually declined after the start of treatment (11,24). Only a few longitudinal studies had used a multi-gene panel for ctDNA analysis (25,26). In a study that utilized a similar gene panel and sequencing method as our study, it was reported that ctDNA reflected tumor load over time. Among 12 patients who had radiographic response to target therapy, their mutant allele concentrations in ctDNA decreased from an average of 10.8% at baseline to 0.2% at a median time of 19 days after treatment initiation. Contrarily, among five patients who did not respond to treatment and had radiographic progressive disease, they showed a relatively high average level of mutant allele fraction at baseline (14.2%) and a modest variation after initiation of therapy (11.8%) were found (26). Our longitudinal study, nevertheless, was more advantageous to demonstrate the utility of ctDNA in monitoring tumor status than previous studies—all our 20 subjects had completed follow-up from baseline, through stable disease, to disease progression, and hence our data showed within-subject variations in ctDNA pattern over the entire course of treatment.
To date, available data support the potential utility of ctDNA as a tumor biomarker for monitoring treatment response and tumor burden over time, particularly when repeat biopsy is not feasible. One question of interest is whether testing for mutations in multiple genes is advantageous over a single gene approach. Our data showed that even in NSCLC patients with a high prevalence of EGFR mutations, testing for multiple genes in ctDNA could help to identify additional patients who had disease progression. Not all mutations detected in ctDNA are actionable or have clinical significance, but changes in ctDNA pattern and quantitative levels informed by a large gene panel are more sensitive in reflecting tumor load than a single gene analysis. Additional benefits of a multi-gene panel include the potential of identifying actionable targets when more drugs are available and revealing therapy resistance mechanisms (9). EGFR mutant patients (Group 3) showed persistent detection of EGFR T790M at Osimertinib baseline through stable disease on treatment. Neither patient had new mutations detected at the time of progression. This observation may imply that tumors bearing acquired EGFR T790M could continue to evolve and may change acquired resistance pathways, through other mechanisms like epithelial-mesenchymal transition (EMT) rather than tumor mutational changes (27).
Different types of TP53 mutations were detectable at different time-points in all the 20 subjects (overall 10/20, 50%), although TP53 mutations appeared to be more prevalent in EGFR wildtype subjects on chemotherapy than in patients treated with EGFR-TKI. TP53 point mutations of various types have been reported to result in loss of tumor suppressor function and promote tumor growth, hence disease progression while on treatment (28,29). The emergence of serial changes in the various different types of TP53 mutations as well as other non-EGFR mutations such as BRCA1, BRCA2, MYC or MET mutations along treatment course probably reflected the evolution of tumor heterogeneity. The contributions of these non-EGFR mutations towards development of acquired drug resistance, regardless of whether the tumor initially carried sensitizing EGFR mutations or not, deserves further investigation (30).
Our study is one of the few in lung cancer literature that assessed serial measurements of ctDNA for a panel of genes. The results provided support for application of serial monitoring of ctDNA mutations in lung cancer patients, especially those on EGFR targeted therapies, and such serial monitoring may allow for discovery of new acquired mutations that could be of therapeutic or prognostication importance. However, this study comprised of a small sample size and lacked a time-to-event outcome, as the level of ctDNA might have started to rise well before clinical progression based on RECIST criteria. As such, we were not able to estimate how much sooner ctDNA could detect disease progression than standard-of-care radiologic imaging, as suggested by some small-scale studies (12,25,26). Future studies need to determine the frequency of ctDNA testing, define the extent of increase in ctDNA, in terms of total quantitative levels or allele fractions of specific genes, to define disease progression, and identify ctDNA biomarkers for early detection of disease progression.
Conclusions
In summary, our results suggest that ctDNA levels in serial blood samples reflect tumor burden over time, and a multi-gene panel would be a more sensitive way of detecting lung cancer disease progression or biomarkers for drug resistance than a single gene approach. The detection of additional oncogenic mutations, some of which are cumulative, or their disappearance in plasma during treatment course, reflects underlying evolution of tumor heterogeneity.
Acknowledgments
The authors thank Ms. Lucinda Wong for data processing and Ms. Peony Chong for data collection and management.
Funding: The research project in this manuscript was supported by the Lee and the Ho Families Respiratory Research Fund.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-675
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-675
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-675). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study protocol was approved by the Ethics Committee of the University of Hong Kong and Hong Kong Hospital Authority Hong Kong West Cluster Institutional Review Board (IRB Reference Number UW 16-104). Informed consent was obtained from all the participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [PubMed]
- Arbour KC, Riely GJ. Systemic therapy for locally advanced and metastatic non-small cell lung cancer: a review. JAMA 2019;322:764-74. [PubMed]
- Zhou J, Kulasinghe A, Bogseth A, et al. Isolation of circulating tumor cells in non-small-cell-lung-cancer patients using a multi-flow microfluidic channel. Microsyst Nanoeng 2019;5:8. [PubMed]
- Kapeleris J, Kulasinghe A, Warkiani ME, et al. The prognostic role of circulating tumor cells (CTCs) in lung cancer. Front Oncol 2018;8:311. [PubMed]
- Kulasinghe A, Kapeleris J, Kimberley R, et al. The prognostic significance of circulating tumor cells in head and neck and non-small-cell lung cancer. Cancer Med 2018;7:5910-9. [PubMed]
- Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [PubMed]
- Lam DC, Tam TC, Lau KM, et al. Plasma EGFR mutation detection associated with survival outcomes in advanced-stage lung cancer. Clin Lung Cancer 2015;16:507-13. [PubMed]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [PubMed]
- Esposito Abate R, Pasquale R, Fenizia F, et al. The role of circulating free DNA in the management of NSCLC. Expert Rev Anticancer Ther 2019;19:19-28. [PubMed]
- Winther-Larsen A, Demuth C, Fledelius J, et al. Correlation between circulating mutant DNA and metabolic tumour burden in advanced non-small cell lung cancer patients. Br J Cancer 2017;117:704-9. [PubMed]
- Imamura F, Uchida J, Kukita Y, et al. Early responses of EGFR circulating tumor DNA to EGFR tyrosine kinase inhibitors in lung cancer treatment. Oncotarget 2016;7:71782-9. [PubMed]
- Taus Á, Camacho L, Rocha P, et al. Dynamics of EGFR mutation load in plasma for prediction of treatment response and disease progression in patients with EGFR-mutant lung adenocarcinoma. Clin Lung Cancer 2018;19:387-94.e2. [PubMed]
- Yanagita M, Redig AJ, Paweletz CP, et al. A prospective evaluation of circulating tumor cells and cell-free DNA in EGFR-mutant non-small cell lung cancer patients treated with erlotinib on a phase II trial. Clin Cancer Res 2016;22:6010-20. [PubMed]
- Mayo-de-Las-Casas C, Garzón Ibáñez M, Jordana-Ariza N, et al. An update on liquid biopsy analysis for diagnostic and monitoring applications in non-small cell lung cancer. Expert Rev Mol Diagn 2018;18:35-45. [PubMed]
- Thompson JC, Yee SS, Troxel AB, et al. Detection of therapeutically targetable driver and resistance mutations in lung cancer patients by next-generation sequencing of cell-free circulating tumor DNA. Clin Cancer Res 2016;22:5772-82. [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [PubMed]
- Phallen J, Sausen M, Adleff V, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med 2017;9:eaan2415. [PubMed]
- Jones S, Anagnostou V, Lytle K, et al. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med 2015;7:283ra53. [PubMed]
- Genovese G, Kähler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 2014;371:2477-87. [PubMed]
- McKerrell T, Park N, Moreno T, et al. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep 2015;10:1239-45. [PubMed]
- Xie M, Lu C, Wang J, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 2014;20:1472-8. [PubMed]
- Zhu YJ, Zhang HB, Liu YH, et al. Quantitative cell-free circulating EGFR mutation concentration is correlated with tumor burden in advanced NSCLC patients. Lung Cancer 2017;109:124-7. [PubMed]
- Karachaliou N, Mayo-de las Casas C, Queralt C, et al. Association of EGFR L858R mutation in circulating free DNA with survival in the EURTAC trial. JAMA Oncol 2015;1:149-57. [PubMed]
- Sorensen BS, Wu L, Wei W, et al. Monitoring of epidermal growth factor receptor tyrosine kinase inhibitor-sensitizing and resistance mutations in the plasma DNA of patients with advanced non-small cell lung cancer during treatment with erlotinib. Cancer 2014;120:3896-901. [PubMed]
- Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov 2017;7:1394-403. [PubMed]
- Phallen J, Leal A, Woodward BD, et al. Early noninvasive detection of response to targeted therapy in non-small cell lung cancer. Cancer Res 2019;79:1204-13. [PubMed]
- Zhang Z, Yang S, Wang Q. Impact of MET alterations on targeted therapy with EGFR-tyrosine kinase inhibitors for EGFR-mutant lung cancer. Biomark Res 2019;7:27. [PubMed]
- Blandino G, Di Agostino S. New therapeutic strategies to treat human cancers expressing mutant p53 proteins. J Exp Clin Cancer Res 2018;37:30. [PubMed]
- Matsumura Y, Owada-Ozaki Y, Suzuki H. Significance of testing for TP53 gene mutations in lung adenocarcinoma using targeted gene sequencing. J Thorac Dis 2018;10:S4147-50. [PubMed]
- Lim ZF, Ma PC. Emerging insights of tumor heterogeneity and drug resistance mechanisms in lung cancer targeted therapy. J Hematol Oncol 2019;12:134. [PubMed]


