
Role of next generation sequencing-based liquid biopsy in advanced non-small cell lung cancer patients treated with immune checkpoint inhibitors: impact of STK11, KRAS and TP53 mutations and co-mutations on outcome
Introduction
The introduction of immune checkpoint inhibitors (ICIs) in clinical practice has revolutionized the treatment of advanced non-small cell lung cancers (NSCLCs) (1-5). Such agents are monoclonal antibodies acting by boosting immune response against cancer cells (6). ICIs commonly used in clinical practice target the PD-1/PD-L1 axis, either inhibiting PD-1 (nivolumab and pembrolizumab) or PD-L1 (atezolizumab and durvalumab). Pembrolizumab is approved as first line treatment for patients with advanced stage NSCLC and tumor cell expression of PD-L1 (tumor proportion score, TPS) ≥50% (4), while the combination of platinum-based chemotherapy plus pembrolizumab is superior to chemotherapy in first-line setting, irrespective of PD-L1 status (7-9). European Medicines Agency (EMA) and Food and Drug Administration (FDA) have also approved atezolizumab in combination with carboplatin and nab-paclitaxel as first line option for non-squamous advanced NSCLCs regardless of PD-L1 expression (9-12). More recently, combinations of nivolumab and ipilimumab, an ICI targeting a different immune checkpoint CTLA-4, and of nivolumab plus ipilimumab and two cycles of platinum-doublet chemotherapy were also approved by FDA for patients with metastatic, non-oncogene driven NSCLC (11,13,14). In pretreated advanced NSCLCs, nivolumab and atezolizumab are approved irrespective of PD-L1 status (1,2,5), whereas pembrolizumab is used only in case of PD-L1 TPS ≥1% (3,9,11).
Immunotherapy has clearly improved the outcome of non-oncogene addicted advanced NSCLC. However, the magnitude of clinical benefit is highly heterogeneous and ICIs might be useless and even detrimental for some patients (15). The detection of PD-L1 TPS via immunohistochemistry has been thus far the most widely studied biomarker for predicting response to ICIs. However, such biomarker has shown a variety of limitations, which impairs its predictive value: heterogeneity of expression, different detection methods, dynamic character (16,17). This is the starting point for the quest for new predictive biomarkers (18). In this field, wide-spectrum mutational analyses performed using high-throughput sequencing tools, such as next-generation sequencing (NGS), have great potentialities. These tools permit relative quantification of tumor genetic alterations as well as their qualitative characterization and may allow for identification of genetic alterations associated with resistance to ICI.
In this regard, the role of alterations of serine-threonine kinase 11 (STK11) gene in NSCLC patients has raised particular interest. STK11 is a tumor suppressor gene coding for a kinase, also known as liver kinase B1 (LKB1), involved in essential cell processes such as metabolic balance, maintenance of DNA integrity and interaction with tissue microenvironment (19-24). This kinase is inactivated in over 30% of lung cancers (25-30) and its impairment seems to be related to an immune desert tumor microenvironment and to a reduced capacity of the transformed cell to recognize DNA damages and stimulate T-cell recruitment (31). In a large retrospective study, tumor genotyping with NGS was performed in tissue and demonstrated that KRAS-mutated non-squamous advanced NSCLC patients carrying STK11 mutations had worse outcome when treated with ICIs, compared to the STK11-wild type counterpart (32). Another study showed that STK11 mutations in advanced NSCLC are associated with lack of durable clinical benefit (i.e., partial response or stable disease lasting more than six months) from immunotherapy (33). It is important to note that these analyses were performed in tissue samples, possibly limiting their applicability in the real-world setting. Indeed, tissue availability for molecular testing is a crucial issue in advanced NSCLC, as sometimes it is even barely enough for histological diagnosis (34-36). In addition, STK11 genetic alterations are widespread in gene and hot-spot testing is not feasible, thus implying the need for relatively high amount of DNA for proper analysis. A possible solution might be the utilization of plasma as source of tumor genetic material.
Aim of the study was to test the role of NGS-based liquid biopsy in advanced NSCLC patients treated with ICIs. In particular, we aimed to confirm the predictive role of STK11 mutations and co-mutations found in plasma in patients treated with ICIs. A parallel control group of patients not receiving ICIs was included.
We present our article in accordance with the STROBE reporting checklist (37) (available at http://dx.doi.org/10.21037/tlcr-20-674).
Methods
Patients
The present study is an observational prospective study, analyzing clinical and molecular data of patients consecutively prospectively screened for two studies: VISION and MAGIC-1.
At our Institution, two trials were active and provided the means to perform NGS testing both in tumor samples and in plasma: the VISION trial (NCT02864992) and the MAGIC (Monitoring Advanced NSCLC through plasma Genotyping during Immunotherapy: Clinical feasibility and application) trial.
The VISION trial is an interventional prospective trial that offered genetic pre-screening with NGS testing of tissue and/or plasma, in order to identify NSCLC patients carrying MET exon 14 skipping alterations or MET amplifications amenable of treatment with tepotinib. Primary endpoints of the trial are activity and tolerability of tepotinib (38,39).
From August 2017 to October 2019, stage IIIB/IV NSCLC patients treated at Rete Oncologica Veneta (ROV) and referring to our Institution for potential eligibility to VISION trial (NCT02864992) were prospectively screened and clinical data were collected.
Main inclusion criteria are: histologically confirmed advanced (Stage IIIB/IV) NSCLC, treatment naïve patients in first-line or pre-treated patients with no more than 2 lines of prior therapy. Main exclusion criteria are: EGFR activating mutations or ALK rearrangements that predict sensitivity to anti-EGFR or anti-ALK therapy respectively, inadequate hematological, renal and hepatic functions and relevant comorbidities, such as impaired cardiac function or known infection with human immunodeficiency virus, or an active infection with hepatitis B or C virus. Pre-screening was performed using NGS testing in tumor tissue and/or in plasma: the choice of the source biological material to be analyzed was at the discretion of the treating physician.
The Ethics Committee of our Institution approved the study on 20th February 2017. The latest version of the protocol was approved by the Ethics committee of our institution on 6th December 2018. A written informed consent was signed before any trial-related activities were carried out. Pre-screening informed consent was obtained prior to NGS testing, either in tumor tissue and/or plasma. Pre-screening informed consent version approved by our Ethics Committee is 3.0. The study was performed in accordance with the Declaration of Helsinki.
The MAGIC study is an observational prospective trial enrolling advanced, non-oncogene addicted NSCLC patients, aimed to identify genetic alterations in tumor tissue, in order to track them down in plasma and subsequently monitor their trend in relation with the kinetic of the disease (40).
From January 2017 and August 2019 advanced NSCLC patients starting systemic treatment at our Institution were prospectively enrolled. Eligibility criteria for MAGIC trial were the absence of known EGFR sensitizing mutations or ALK or ROS-1 rearrangements, availability of tumor biopsy material collected before starting any treatment, the planning of systemic treatment and the possibility of adequate clinical and radiological follow-up. Patients were treated according to standard clinical practice with chemotherapy or ICIs. Main exclusion criteria were chemo- or radiation treatments performed prior to tissue collection. According to clinical practice, radiological evaluation was performed with iodine contrast CT-scan at baseline and during treatment.
The Ethics Committee of our Institution evaluated and approved study design and informed consent (2016/82, on 12th December 2016). Written informed consent was obtained from all patients before study entry. The study was performed in accordance with the Declaration of Helsinki.
The aim of this study (LINE, LKB1 and Immune involvement in advanced NSCLC: an Exploratory analysis) was to genetically characterize advanced NSCLC patients and in particular to define the role of LKB1 in patients’ outcome while on ICI. For this purpose, patients enrolled in the MAGIC and VISION trials were considered as study population, if tested in plasma before the start of ICIs, and as control group, if never treated with ICIs. Patients tested during the course of immunotherapy were excluded. Patients’ data recorded at baseline included patient demographics, Eastern Cooperative Oncology Group (ECOG) performance status (PS) at time of first line systemic treatment start, smoking history and weigh loss of more than 5% during 6 months before cancer diagnosis. Tumor data collected included histology, EGFR, ALK and ROS-1 status, as they were determined for non-squamous NSCLC, PD-L1 status, when available, and radiological staging. Information about treatments undergone by patients during the course of the disease, their response and toxicity were also collected.
The Ethics Committee of our Institution evaluated and approved this study and informed consent on 22nd January 2018 (2018/21). Written informed consent was obtained from all patients before study entry. The study was performed in accordance with the Declaration of Helsinki.
Molecular analyses
In the frame of VISION trial, circulating tumor DNA (ctDNA) was isolated and tested from freshly collected plasma samples. ctDNA (5.0 to 30 ng) was extracted, enriched for targeted regions and underwent library preparation, in order to complete sequencing to be conducted using Illumina platform. Tumor tissue for NGS testing was obtained from archived samples or from freshly obtained formalin-fixed paraffin-embedded (FFPE) tumor tissue. MET alterations were searched in either plasma samples or tissue tumor samples, using Guardant360® Test, covering 73 genes and all somatic alterations recognized as potential targets by NCCN, and Oncomine™ Focus Assay (testing done by MolecularMD Inc.) covering 59 genes, respectively. Further details are described in Supplementary file (Appendix 1).
ctDNA of patients participating in MAGIC trial was isolated and tested from freshly collected plasma samples. Tumor tissue for NGS testing was obtained from archived samples or from freshly obtained FFPE tumor tissue. Plasma samples were collected at the time of first administration of systemic treatment (baseline, T1), after three or four weeks of treatment (according to the treatment schedule) (3±1 weeks, T2), and at first radiological restaging (T3). For the present study, genetic testing performed at baseline was evaluated. ctDNA was extracted from 3 to 5 mL of plasma using the QIAamp® circulating nucleic acid Kit (QIAGEN, Hilden, Germany). CtDNA (2 to 25 ng) was used for library preparation, following the protocol of Myriapod NGS-IL 56G (Diatech Pharmacogenetics Srl; Jesi, Italy) assay. This assay covers mutational hotspots of 56 tumor-associated genes. Further details about study procedures are described in Supplementary file (Appendix 1).
Statistical analyses
The primary aim of the study was to describe the role of plasma NGS in characterizing advanced NSCLC patients receiving ICIs according to clinical practice. Primary end-point was to depict the association of the presence of the most frequent tumor-associated genetic alterations with outcome.
We analyzed the frequency of genetic alterations found and the association of the number of genetic alterations found with clinical features. Subsequently, we evaluated the impact of the presence of the most frequent genetic alterations, i.e., the ones exceeding median number of alterations found per gene, with outcome. For patients enrolled in both trials and with NGS results available from Guardant360® Test and Myriapod NGS-IL 56G Assay, we chose to use the results from the former gene panel, given its width of genome coverage. Gene variants were searched in the COSMIC database and pathogenicity was defined accordingly.
Outcome was considered in terms of overall survival (OS), as primary end-point, of immune-related progression free survival (irPFS) and of radiological response. OS was calculated from the first day of any systemic treatment to death from any cause. IrPFS was calculated from the first day of ICI treatment to the first radiological or clinical disease progression or death from any cause. Response was assessed according to Response Evaluation Criteria in Solid Tumors (RECIST) v1.1; disease control rate (DCR) was defined as complete response (CR) plus partial response (PR) plus stable disease (SD).
Variables were presented by using median value for continuous variables and percentages (numbers) for categorical variables and their relationship with the presence of target alteration was assessed using Mann-Whitney test, Kruskal-Wallis test and the chi-squared test as appropriate. Correlation among different gene alterations was calculated using Pearson’s correlation test.
Univariate logistic regression models and results were reported using odds ratio (OR) with their 95% confidence interval (CI). Median PFS and OS were estimated by using Kaplan-Meier methods and the log-rank test was used to compare survival between groups. Hazard ratios (HR) and their 95% CI were calculated with the Cox regression method. Potential confounding effect of clinical variables was assessed by performing multivariate analysis with Cox regression method.
All the analyses were also performed in a control group of patients not receiving ICIs.
Statistical significance level was set at P<0.05 for all tests. All statistical analyses were performed with statistical package for the social sciences (SPSS) 20.0 software (SPSS Inc., Chicago, IL, USA).
Results
Patients and treatments
Among 234 NSCLC patients prospectively screened with NGS in plasma, 103 patients received ICIs for advanced disease and had their plasma collected right before ICIs initiation. Among them, 72 had been pre-screened in VISION trial, 79 in the MAGIC trial; 48 of them participated in both studies. All plasma samples resulted evaluable.
Gender distribution was: 64 (62.1%) men and 39 (37.9%) women. Seventeen patients (16.5%) were never smokers, whereas 86 (83.5%) were either former (n=41; 39.8%) or active smokers (n=45; 43.7%). Median age at diagnosis of metastatic disease was 69.3 (range, 42.1–84.9) years. At the time of advanced NSCLC diagnosis, 26 (25.2%) subjects had an ECOG PS of 0 and 77 (74.8%) of ≥1. Nineteen patients (18.4%) experienced significant weight loss prior to NSCLC diagnosis. At the time of diagnosis of stage IV NSCLC, 39 subjects (37.9%) had one single site of metastasis, 39 (37.9%) had two and 25 (24.2%) had three or more sites of distant disease. Fifty-two (50.5%) patients had extra-thoracic sites of metastasis, among them 15 (14.6%) had liver metastases and 29 (28.2%) had bone metastases (Table 1).

Full table
Most of the patients were diagnosed with adenocarcinoma (n=80; 77.7%); the others were squamous cell carcinomas (n=20; 19.4%), non-otherwise specified carcinomas (n=2; 1.9%), or sarcomatoid carcinomas (n=1; 1.0%). PD-L1 status was available for 95 patients (92.2%). Using a 1% cut-off for PD-L1 TPS, 45 cases (47.4%) were positive. Thirty-one patients (32.6%) had PD-L1 TPS ≥50%.
For advanced disease, 19 patients (18.4%) received only one line of treatment, 66 (64.1%) two lines and 18 (17.5%) at least three lines. Immunotherapy was administered as first line treatment in 27 cases (26.2%).
A control group of 101 patients never underwent ICI treatment and had their plasma collected. Most of them were men (n=57, 56.4%) and former or current smokers (n=73, 72.3%). At the time of diagnosis of stage IV NSCLC, 31 (30.7%) had an ECOG PS 0 and 70 (69.3%) had an ECOG PS equal or higher than 1. Adenocarcinoma was the most frequent histotype (n=80, 79.2%). At the time of diagnosis of advanced disease, 49 subjects (48.5%) had one single site of metastasis, 33 (32.7%) had two and 19 (18.8%) had three or more sites of distant disease.
Clinical and pathological characteristics are displayed in Table 1.
Response to ICIs and patients’ outcome
At the time of the analysis, median follow-up time was 20.3 (range, 5.1–57.8) months. DCR was 54.4% (partial response n=14, 13.6%, and stable disease n=42, 40.7%). Median OS was 20.8 (95% CI: 16.7–24.9) months. Median irPFS was 4.2 (95% CI: 2.3–6.1) months.
Impact of clinical features on patients’ outcome
We performed univariate analysis in order to evaluate the impact on OS of known clinical prognostic features. ECOG PS (HR =1.775, 95% CI: 1.101–2.862, P=0.019), non-adenocarcinoma histology (HR =2.158, 95% CI: 1.158–4.021, P=0.015), having extrathoracic metastases (HR =2.600, 95% CI: 1.480–4.567, P=0.001) and bone metastases (HR =2.137, 95% CI: 1.197–3.816, P=0.010) had a significant impact on OS. In multivariate analysis, ECOG PS (HR =2.158, 95% CI: 1.108–4.205, P=0.024) and the presence of extrathoracic lesions (HR =2.932, 95% CI: 1.287–6.681; P=0.010) were confirmed to impact OS independently (Table 2).
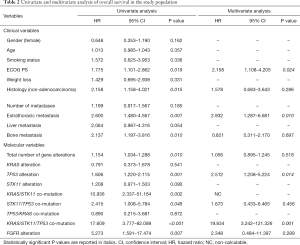
Full table
The same variables were tested for impact on irPFS. ECOG PS (HR =2.053, 95% CI: 1.266–3.331, P=0.004) and having extrathoracic metastases (HR =2.054, 95% CI: 1.272–3.316, P=0.003) or bone metastases (HR =1.847, 95% CI: 1.120-3.046, P=0.016) had significant impact on irPFS. When multivariate analysis was performed, significance was maintained for ECOG PS (HR =2.692, 95% CI: 1.488–4.872, P=0.001) and for the presence of extrathoracic metastases (HR =2.417, 95% CI: 1.154–5.065, P=0.019) (Table 3).
Full table
Genetic alterations in ctDNA
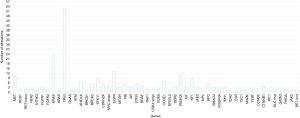
The median number of mutations detected in a patient was 2 (range, 0–10). In 21 cases (20.4%) no mutation was detected. Genetic alterations were detected in fifty-three genes and among them the median number of alteration for each gene was 3 (range, 1–53) (Figure 1). The most commonly altered genes were: TP53 (n=53), KRAS (n=22), EGFR (n=13), PIK3CA (n=11), MET (n=10), STK11 (n=9), NF1 (n=9), CCNE1 (n=7), ARID1A (n=6), BRCA2 (n=5), RB1 (n=5), CDKN2A (n=5), PTEN (n=5), BRAF (n=5), APC (n=5), MYC (n=4), CTNNB1 (n=4), MTOR (n=3), FGFR1 (n=3), AR (n=3), DDR2 (n=3), KIT (n=3), SMAD4 (n=3), PDGFRA (n=3) and IDH1 (n=3).
TP53 genetic alterations were detected in 53 cases (51.5%) and nine patients carried more than one TP53 pathogenic variant. Among these 53 cases, two carried a variant of unknown significance (VUS) and two carried a neutral variant (Functional Analysis through Hidden Markov Models, FATHMM, score of 0.24); the other 49 were either pathogenic or likely pathogenic (Table S1).
KRAS mutations were found in 22 cases (21.3%), with one patient only carrying a VUS mutation. Most of these mutations (n=16, 72.7%) caused a change in the amino acid residue at position 12 (G12X). EGFR was mutated in nine cases (8.7%); four alterations were VUS, the others were non-classical mutations. EGFR gene was amplified in four cases. Nine patients carried PIK3CA mutations: seven had a pathogenic alteration (FATHMM between 0.96 and 1.00), while two had a benign polymorphism. Furthermore, PIK3CA was amplified in two more cases. Three patients carried a MET exon 14 skipping mutation; two of these carried also a low MET gene amplification. Two patients carried other pathogenic MET mutations, two carried neutral MET mutations, one had a VUS of MET and two had a MET amplification. NF1 was also altered in nine cases (9.7%) and six of them were defined as VUS, while the others were pathogenic with a FATHMM score ranging between 0.93 and 0.99.
STK11 gene alterations were detected in nine samples (8.7%) and they were all pathogenic variants. The most common alterations were nonsense mutations (n=4, 44.5%), causing premature stop codons, followed by pathogenic missense mutations (n=3, 33.3%) and splice site single nucleotide variants (n=2, 22.2%). Three patients (2.9%) carried concomitant KRAS/STK11 gene alterations, of whom two patients (2.2%) carried also a concomitant TP53 alteration. Moreover, five patients (4.8%) carried a TP53 alteration with a STK11 mutation.
Details about the other gene alterations are displayed in Table S2.
TP53, KRAS and STK11 alterations and clinical features
Among clinical and histological features, being a former or current smoker was associated with the presence of TP53 pathogenic alterations (OR =4.020, 95% CI: 1.211–13.399, P=0.023) (Figure 2A). In addition, carrying TP53 alterations was correlated with higher risk of having extrathoracic metastases (OR =1.703, 95% CI: 1.135–2.556, P=0.010) (Figure 2B).
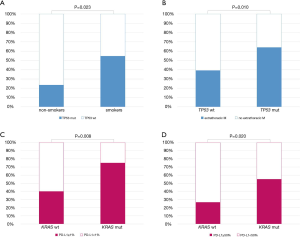
KRAS mutations were not significantly associated with any clinical features and neither were STK11 genetic alterations.
Moreover, the presence of KRAS mutations was associated with PD-L1 expression in tumor cells, both using 1% (OR =4.500, 95% CI: 1.479–13.690, P=0.008) and 50% (OR =3.361, 95% CI: 1.213–9.310, P=0.020) as cut-off (Figure 2C,D). Table S3 summarizes the impact of clinical features on the presence of genetic alterations.
Association between different gene alterations
We also evaluated the presence of any association between gene alterations found in our study population. As shown in Figure 3, CDKN2A, MTOR, STK11, CTNNB1, RB1 and APC alterations were the ones with the highest number of statistically significant associations with other gene alterations.
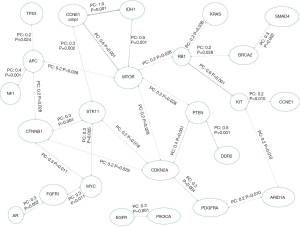
In particular, the presence of a mutated CDKN2A was positively associated with a mutated CTNNB1 (Pearson’s correlation coefficient, PC, of 0.2, P=0.029), STK11 (PC=0.2, P=0.018), MTOR (PC=0.2, P=0.029), PTEN (PC=0.4, P=0.001) and PDGFRA (PC=0.3, P=0.004). MTOR mutations were associated also with APC mutations (PC=0.2, P=0.028), CCNE1 amplification (PC=0.6, P=0.001), IDH1 mutation (PC=0.5, P=0.001), a mutated RB1 (PC=0.2, P=0.028) and a mutated PTEN (PC=0.2, P=0.028).
A mutated STK11, in turn, was linked also with CTNNB1 mutation (PC=0.3, P=0.004), MYC amplification (PC=0.3, P=0.005) and CCNE1 amplification (PC=0.3, P=0.002).
Besides STK11 and CDKN2A, CTNNB1 mutations were associated with APC mutations (PC=0.2, P=0.028) and MYC amplifications (PC=0.3, P=0.011). Moreover APC alterations were also linked with NF1 (PC=0.4, P=0.001) and with TP53 mutations (PC=0.2, P=0.024), while a mutated RB1 was also associated with KRAS (PC=0.2, P=0.038), BRCA2 (PC=0.2, P=0.028) and KIT mutations (PC=0.6, P=0.001). With regards of TP53 and KRAS mutations, they were significantly associated with APC (PC=0.2, P=0.024) and RB1 (PC=0.2, P=0.038) alterations, respectively.
Details about other genes and their mutual correlations are depicted in Figure 3.
Impact of genetic alterations on treatment response and patients’ outcome
When we considered the total number of gene alterations detected in plasma, we found that a higher number of genetic alterations was correlated with shorter OS (HR =1.154, 95% CI: 1.034–1.288, P=0.010), but it did not affect irPFS or DCR.
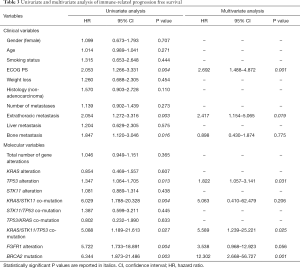
Patients carrying pathogenic TP53 alterations had significantly shorter median OS, when compared to the TP53 wildtype counterpart (12.6 months, 95% CI: 8.5–16.8, vs. 26.7, 95% CI: 17.4–36.1; HR =1.606, 95% CI: 1.220–2.115, P=0.001) (Figure 4). TP53 affected negatively also irPFS (3.2 months, 95% CI: 1.6-4.8 vs. 6.6 months, 95% CI: 1.9–11.3; HR =1.347, 95% CI: 1.064–1.705, P=0.013) (Table 2). DCR was not affected by TP53 status (OR =0.745, 95% CI: 0.504–1.103, P=0.142).
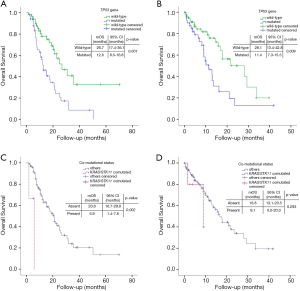
The presence of STK11 alterations was associated with a trend for shorter OS (median OS: 14.9 months, 95% CI: 10.5–19.3, vs. 21.2 months, 95% CI: 17.3–25.1; HR =1.208, 95% CI: 0.971-1.503, P=0.090), although not reaching statistical significance. STK11 mutational status did not affect irPFS (2.4 months, 95% CI: 2.0–2.8, vs. 4.4 months, 95% CI: 2.7-6.0; HR =1.081, 95% CI: 0.889–1.314, P=0.438) (Table 2) or DCR (OR =0.789, 95% CI: 0.550–1.132, P=0.197).
The presence of KRAS mutation did not significantly affect OS, irPFS and DCR (Table 2). This was confirmed also when we analyzed G12X mutations versus non-G12X mutations. Of interest, patients with KRAS G12X amino-acid substitution had a trend for better OS compared with non-G12X amino-acid substitution, even though it was not statistically significant (35.6 months, 95% CI: 8.1–62.9, vs. 21.2 months, 95 CI%: non-evaluable, P=0.732, log-rank test).
The presence of other alterations in previously mentioned genes did not significantly affect patients’ outcome (data not shown), with the exception of FGFR1 whose alterations had a negative impact both on OS (HR =5.722, 95% CI: 1.733–18.891, P=0.004) and on irPFS (HR =5.273, 95% CI: 1.591–17.474, P=0.007), and of BRCA2, which affected irPFS (HR =6.344, 95% CI: 1.873–21.486, P=0.003).
We also evaluated the impact of the presence of KRAS/STK11 co-mutation (Figure 4, Tables 2,3). This status was associated both with worse OS (5.9 months, 95% CI: 1.4–7.6, vs. 20.8 months, 95% CI: 16.7–29.9; HR =10.936, 95% CI: 2.337–51.164, P=0.002) and worse irPFS (1.2 months, 95% CI: 0.9–1.5, vs. 4.4 months, 95% CI: 2.7–6.0, HR =6.029, 95% CI: 1.788–20.328, P=0.004). STK11/TP53 co-mutation had impact on OS (14.9 months, 95% CI: 10.3–19.5, vs. 21.2 months, 95% CI: 17.4–25.0; HR =2.415, 95% CI: 1.008–5.784, P=0.048), but not on irPFS (2.4 months, 95% CI: 2.1–2.8, vs. 4.4 months, 95% CI: 2.7–6.0; HR =1.387, 95% CI: 0.599–3.211, P=0.445). The presence of TP53/KRAS co-mutation did not impact either OS or irPFS. On the contrary, the presence of KRAS/STK11/TP53 concomitant mutations negatively affected OS (1.8 months, 95% CI: not calculable, vs. 20.8, 95% CI: 16.9–24.8; HR =17.609, 95% CI: 3.777–82.089, P<0.001) and irPFS (1.0 month, 95% CI: not calculable, vs. 4.4, 95% CI: 2.7-6.1; HR =5.088, 95% CI: 1.189–21.613, P=0.027).
We performed multivariate OS analysis including ECOG PS, histology, the presence of extrathoracic or bone metastases, total number of gene alterations, TP53 mutation, KRAS/STK11, STK11/TP53 or KRAS/STK11/TP53 co-mutation and FGFR1 alteration as covariates. TP53 and KRAS/STK11/TP53 co-mutation confirmed their independent impact on OS (HR =2.512, 95% CI: 1.208–5.224, P=0.014 and HR =19.834, 95% CI: 3.242–121.326, P=0.001, respectively) (Table 2).
Finally, we performed multivariate irPFS analysis including ECOG PS, the presence of extrathoracic or bone metastases, TP53 alterations, KRAS/STK11 or KRAS/STK11/TP53 co-mutation, FGFR1 alteration and BRCA2 mutation. Negative impact on patients’ irPFS of TP53, KRAS/STK11/TP53 co-mutation and BRCA2 mutation was confirmed (HR =1.822, 95% CI: 1.057–3.141, P=0.031; HR =5.589, 95% CI: 1.239–25.221, P=0.025; HR =12.302, 95% CI: 2.668–56.727, P=0.001) (Table 3).
Genetic characterization and outcome in a control group of patients never treated with ICIs
A control population of advanced NSCLC patients (n=101) who never received immunotherapy was also evaluated. Patients’ characteristics were well balanced between populations treated or not with ICIs, with the exception of enrichment in PD-L1-strong-positive (TPS ≥50%) among patients treated with ICIs (Table 1). At the time of the analysis, median follow-up time was 13.2 (range, 1.8–43.0) months for the control group. Median OS of control population was 16.8 months (95% CI: 13.0–20.6).
In univariate analysis, ECOG PS (HR =2.310, 95% CI: 1.222–4.365, P=0.010), weight loss (HR =4.133, 95% CI: 2.169–7.875, P<0.001), number of metastases (HR =1.703, 95% CI: 1.268–2.286, P<0.001) and having extrathoracic metastases (HR =1.958, 95% CI: 1.035–3.701, P=0.039) or bone metastasis (HR =3.181, 95% CI: 1.689–5.991, P<0.001) had significant impact on OS. Weight loss (HR =3.499, 95% CI: 1.766–6.933, P<0.001) and the presence of bone metastases (HR =3.179, 95% CI: 1.366–7.399, P=0.007) maintained independent prognostic effect in multivariate analysis.
Analyzing the impact of mutational status, TP53 and KRAS mutations had a negative impact on OS (HR =1.498, 95% CI: 1.105–2.029, P=0.009, Figure 4B, and HR =2.999, 95% CI: 1.525–5.899, P=0.001 respectively), confirmed in multivariate test (Table S4), while STK11 had no impact at all. Neither STK11/KRAS, nor STK11/KRAS/TP53, nor STK11/TP53 co-mutation had any impact on patients’ outcome (HR =2.180, 95% CI: 0.510–9.326, P=0.293, HR =2.411, 95% CI: 0.324–17.913, P=0.390 and HR =2.051, 95% CI: 0.628–6.698, P=0.234, respectively) (Figure 4). When tested in multivariate analysis, both TP53 and KRAS alterations were still significantly associated with worse survival (HR =1.422, 95% CI: 1.045–1.936, P=0.025 and HR =2.701, 95% CI: 1.362–5.356, P=0.004, respectively; Table S4).
Discussion
Wide genetic characterization has the potential to provide predictive information for patients treated with ICIs, but tissue availability is often a limitation that may impair its application in clinical practice.
In the present work, we characterized patients undergoing immunotherapy, using NGS testing in liquid biopsies performed before the first ICI administration, in order to define the impact of genetic alterations found in plasma on outcome.
Since tissue availability often represents a strong limitation to wide molecular characterization, the use of plasma as the source of ctDNA is surely promising: liquid biopsy is a feasible procedure and could be easily repeated. ctDNA may also be representative of tumor burden and mirrors intrinsic tumor heterogeneity in space and time (40-45).
In our series, liquid biopsy samples were evaluable in the whole cohort and TP53 mutation was the most frequent gene alteration, carrying also prognostic value. The presence of TP53 mutations was associated with shorter OS both in patients receiving ICIs and in patients who never received immunotherapy. The prognostic role of TP53 in lung cancer is still debated. It has been studied extensively in surgical series and a meta-analysis confirmed its prognostic value in early stage disease only in adenocarcinoma histological subtype (46). In the studies included in this meta-analysis, however, samples were not tested using a high-throughput sequencing technique, such as NGS. More recently, La Fleur and colleagues used NGS to describe a significant negative impact of TP53 alterations (HR =1.47, P=0.003) in a cohort of NSCLC treated with surgery (47). In the setting of advanced disease, a large report on over 1,400 metastatic patients showed similar results and median OS of TP53 mutated was 8 months shorter than the wild-type counterpart (48). Other reports did not confirm significant prognostic value of TP53 mutations in advanced NSCLC (49-51). To date, to the best of our knowledge, ours is the first report describing such correlation using only plasma as source material for NGS.
In our ICI cohort, STK11 mutations were identified in 9% of patients, in line with previous reports (range: 8% to 13%) (47,50-52). Patients carrying a pathogenic variant of STK11 and receiving ICIs showed a trend for shorter OS. The prognostic role of STK11 has been already investigated in tissue samples, and previous retrospective studies found a negative impact on outcome (50,52). These studies included patients treated with first-line platinum-based chemotherapy, with no details about further lines of treatment. Moreover, one of the two studies was characterized by a median follow-up time of only 7.39 months, leading to a median OS of 6.68 months (50), which looks even shorter than pre-immunotherapy historical controls (53). Furthermore, a recent post-hoc analysis by Vernieri and colleagues questioned the clinical impact of STK11 mutations in NSCLC patients treated chemotherapy in the frame of the TAILOR trial (54). Therefore, any comparison should be made with caution. On the other hand, tissue NGS analysis in 240 patients treated with anti-PD-1/PD-L1 agents showed that STK11 mutated status was significantly enriched in the subgroup of patients who did not obtain a durable clinical benefit, defined as not progressive disease for at least 6 months (33,55). Similarly, Guibert et al. evaluated 97 patients treated with ICIs using a targeted plasma NGS at treatment baseline and found that STK11-mutated patients derived a shorter irPFS, compared with the wild-type ones (51).
In our series, we were able to investigate the role of specific co-occurring gene mutations in modulating STK11 function. First, we reported how KRAS/STK11 co-mutations had a negative impact both on OS and on irPFS. No significant impact on outcome was found among patients not receiving ICIs.
The predictive role for KRAS/STK11 co-mutations detected in tissue in patients receiving ICIs was recently depicted by Skoulidis et al. (56). The study included a control group of 120 patients treated with chemotherapy and showing no impact of co-mutations on outcome (56). On the other hand, potential negative prognostic impact of KRAS/STK11 co-mutations in tissue emerged in previous studies, in which study population was heterogeneous according to treatment (47,52,57). In particular, a recent work selected a population of advanced NSCLC patients carrying STK11 alterations (n=62) using tissue-based (n=44) or plasma-based (n=18) NGS, in order to define the impact of concomitant alterations on prognosis and confirmed the negative role of KRAS/STK11 co-mutations in the study population, also including patients treated with first-line ICI (58). Notably, in our series we were able to describe differential impact on prognosis of KRAS/STK11 co-mutation, according to the fact of being treated or not with ICIs. This finding confirms potential predictive role of KRAS/STK11 co-mutations in patients treated with ICIs and validate plasma NGS testing for the identification of such alterations.
KRAS mutations are frequent alterations in NSCLC and KRAS-mutated disease represents a heterogeneous group. The subtype of KRAS mutation affects prognostic impact (59) and potentially also therapeutic options (60,61), while the presence of co-mutations modulates its cellular functions. In particular, the cross-talk between the KRAS-MAPK signalling pathway and the one of LKB1-AMPK has been investigated (62). The oncogenic path primed by KRAS mutation is further boosted by LKB1 inactivation, which is able to increase metastatic potentials acting on the anchorage-dependent cell-growth (23) and tumor cell differentiation (63). LKB1 inactivation in KRAS-mutated tumors also impairs one of the first steps of the immune-surveillance process, through the inhibition of stimulator of interferon genes (STING) pathway (64), leading to an immune-excluded tumor immune micro-environment (65).
Even though the number of patients carrying co-mutations is limited, we also depicted the impact of concomitant KRAS/STK11/TP53 mutations on outcome, according to treatment. In this context, Bange and colleagues did not find any significant impact of KRAS/STK11/TP53 co-mutations on outcome. The study population was heterogeneous according to treatment and the timing of liquid biopsy was not pre-defined and could occur over-90 days after the start of first line therapy (58).
In ICI cohort of our study, also FGFR1 turned out to have negative impact on patients’ OS and irPFS, even if it was not confirmed in multivariate analysis (Tables 2,3). In this field, evidence is very scanty. FGFR1 was included in the gene panel tested in the previous series of ICI-treated patients, but no impact on outcome was reported (33,51). As shown in preclinical experiences with breast cancer cell-lines, a dysregulated FGFR1 pathway could induce an immune-suppressive microenvironment, through macrophage recruitment and increased angiogenesis (66-68). However, larger studies are needed to evaluate and eventually validate this finding in NSCLC field.
We also found that BRCA2 pathogenic mutations negatively affected irPFS. Evidence about the impact of this alteration in lung cancer is scarce. Experiences in other neoplasms, such as breast, ovarian and prostate cancers, seem to correlate the presence of mutations affecting BRCA2 to higher mutational burden and to a STING-mediated activation of the immune system (69), but also to an immune-suppressive tumor microenvironment (70) and to the absence of activated T-cells (71). Given to the low number of BRCA2 mutated patients in our series, we are not able to draw any conclusion; however, the role of BRCA2 alterations remains controversial and needs to be further evaluated in NSCLC patients, in particular when immunotherapy is administered.
Among the other most common alterations, we detected nine non-classical EGFR mutations (Table S2). EGFR common alterations (i.e., exon 19 deletions and exon 21 mutations) are associated with reduced efficacy of immunotherapy, but information about the impact of rare EGFR alterations on immunotherapy is unknown (72). In our series, the presence of non-classical EGFR mutations did not significantly affect patients’ outcome.
The main strengths of our study are the consecutive enrolment of patients referred to our center, the extensive performance of liquid biopsy in a real-world population and the possibility to analyze the results according to treatment. Our control group was adequate in terms of number of patients and of clinical and pathological homogeneity with study population. NGS multi-gene screening let us also establish the role of co-mutations, whose role in modulating oncogenic pathways is known and deserves further clinical validation (32,73-75). As a matter of fact, when analyzing KRAS, STK11 and TP53 mutations and co-mutations according to treatment, the predictive value of STK11 co-mutation clearly emerged.
The present study has also some limitations. First, when analyzing STK11 status, we could technically evaluate point mutations (SNV) and small insertions and deletions, but not large deletions involving the STK11 gene, nor its epigenetic modifications, that can also account for STK11 inactivation (30,32,76). Therefore, our conclusions may underestimate the real impact of altered STK11 on patients’ response to immunotherapy. Second, the total number of STK11-mutated cases or carrying other co-mutations was limited. Third, regarding the NGS gene panel used to test plasma samples in our series, other important gene alterations, such the ones affecting KEAP1, were not included. The KEAP1 pathway has a central role in protecting cells from oxidative and electrophilic stresses (77) and is disrupted in about 15% of NSCLC (78). KEAP1 mutational status and its epigenetic silencing are associated with poor prognosis and chemotherapeutic resistance in NSCLC (79). Furthermore, it seems to be also linked to an “immune-cold” tumor microenvironment (80,81) and, when present with a concurrent KRAS mutation, it was associated with shorter irPFS (57).
Conclusions
Our study depicts the role of liquid biopsy in detecting tumor genetic alterations with potential predictive impact in patients undergoing immunotherapy in clinical practice. While TP53 mutations showed to have prognostic value, we confirmed the predictive value of KRAS/STK11 and KRAS/LKB1/TP53 mutations when detected in plasma.
Acknowledgments
Funding: The molecular analyses were performed for the VISION trial and they were all provided by Merck. The consent for publication of the molecular data and correlation with clinical features and outcome has been provided by Merck Medical Direction. Merck was allowed to review the manuscript but was not involved in this project more than providing the molecular analyses as part of the screening for the VISION trial. This work was supported by IOV intramural research grant 2017 – SINERGIA (to LB) and IOV intramural research grant 2019 (to SI).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-674
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-674
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-674). RR serves as a current Editor-in-chief for Translational Lung Cancer Research. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The Ethics Committee of our Institution evaluated and approved the VISION study and informed consent on 20th February 2017 (2017/52). A written informed consent was signed before any trial-related activities were carried out. The Ethics Committee of our Institution approved MAGIC study design and informed consent on 12th December 2016 (2016/82). A written informed consent was obtained from all patients before study entry. The Ethics Committee of our Institution evaluated and approved LINE study and informed consent on 22nd January 2018 (2018/21). Written informed consent was obtained from all patients before study entry. All three studies were performed in accordance with the Declaration of Helsinki.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:1627-39. [Crossref] [PubMed]
- Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:123-35. [Crossref] [PubMed]
- Herbst RS, Baas P, Kim DW, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387:1540-50. [Crossref] [PubMed]
- Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2016;375:1823-33. [Crossref] [PubMed]
- Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 2017;389:255-65. [Crossref] [PubMed]
- Raju S, Joseph R, Sehgal S. Review of checkpoint immunotherapy for the management of non-small cell lung cancer. ImmunoTargets Ther 2018;7:63-75. [Crossref] [PubMed]
- Gandhi L, Rodríguez-Abreu D, Gadgeel S, et al. Pembrolizumab plus Chemotherapy in Metastatic Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:2078-92. [Crossref] [PubMed]
- Paz-Ares L, Luft A, Vicente D, et al. Pembrolizumab plus Chemotherapy for Squamous Non-Small-Cell Lung Cancer. N Engl J Med 2018;379:2040-51. [Crossref] [PubMed]
- Planchard D, Popat S, Kerr K, et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann Oncol 2018;29:iv192-237. [Crossref]
- West H, McCleod M, Hussein M, et al. Atezolizumab in combination with carboplatin plus nab-paclitaxel chemotherapy compared with chemotherapy alone as first-line treatment for metastatic non-squamous non-small-cell lung cancer (IMpower130): a multicentre, randomised, open-label, phase 3 tria. Lancet Oncol 2019;20:924-37. [Crossref] [PubMed]
- Ettinger DS, Wood DE, Aisner DL, et al. Non-Small Cell Lung Cancer - National Comprehensive Cancer Network (NCCN) - 2020 Clinical Practice Guidelines - Version 8.2020. 2020.
- Hanna NH, Schneider BJ, Temin S, et al. Therapy for Stage IV Non-Small-Cell Lung Cancer Without Driver Alterations: ASCO and OH (CCO) Joint Guideline Update. J Clin Oncol 2020;38:1608-32. [Crossref] [PubMed]
- Reck M, Ciuleanu TE, Dols MC, et al. Nivolumab (NIVO) + ipilimumab (IPI) + 2 cycles of platinum-doublet chemotherapy (chemo) vs 4 cycles chemo as first-line (1L) treatment (tx) for stage IV/recurrent non-small cell lung cancer (NSCLC): CheckMate 9LA. J Clin Oncol 2020;38:abstr 9501.
- Hellmann MD, Paz-Ares L, Bernabe Caro R, et al. Nivolumab plus Ipilimumab in Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2019;381:2020-31. [Crossref] [PubMed]
- Ferrara R, Mezquita L, Texier M, et al. Hyperprogressive Disease in Patients with Advanced Non-Small Cell Lung Cancer Treated with PD-1/PD-L1 Inhibitors or with Single-Agent Chemotherapy. JAMA Oncol 2018;4:1543-52. [Crossref] [PubMed]
- Bai R, Lv Z, Xu D, et al. Predictive biomarkers for cancer immunotherapy with immune checkpoint inhibitors. Biomark Res 2020;8:34. [Crossref] [PubMed]
- You W, Shang B, Sun J, et al. Mechanistic insight of predictive biomarkers for antitumor PD-1/PD-L1 blockade: A paradigm shift towards immunome evaluation Oncol Rep 2020;44:424-37. (Review). [Crossref] [PubMed]
- Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer 2019;19:133-50. [Crossref] [PubMed]
- Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: Metabolism and growth control in tumour suppression. Nat Rev Cancer 2009;9:563-75. [Crossref] [PubMed]
- Lizcano JM, Göransson O, Toth R, et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J 2004;23:833-43. [Crossref] [PubMed]
- Wang Y-S, Chen J, Cui F, et al. LKB1 is a DNA damage response protein that regulates cellular sensitivity to PARP inhibitors. Oncotarget 2016;7:73389-401. [Crossref] [PubMed]
- Liang X, Wang P, Gao Q, et al. Exogenous activation of LKB1/AMPK signaling induces G1 arrest in cells with endogenous LKB1 expression. Mol Med Rep 2014;9:1019-24. [Crossref] [PubMed]
- Hezel AF, Bardeesy N. LKB1; linking cell structure and tumor suppression. Oncogene 2008;27:6908-19. [Crossref] [PubMed]
- Bonanno L, Zulato E, Pavan A, et al. LKB1 and Tumor Metabolism: The Interplay of Immune and Angiogenic Microenvironment in Lung Cancer. Int J Mol Sci 2019;20:1874. [Crossref] [PubMed]
- Wingo SN, Gallardo TD, Akbay EA, et al. Somatic LKB1 Mutations Promote Cervical Cancer Progression. Aziz SA, editor. PLoS One 2009;4:e5137.
- Tanwar PS, Mohapatra G, Chiang S, et al. Loss of LKB1 and PTEN tumor suppressor genes in the ovarian surface epithelium induces papillary serous ovarian cancer. Carcinogenesis 2014;35:546-53. [Crossref] [PubMed]
- Morton JP, Jamieson NB, Karim SA, et al. LKB1 Haploinsufficiency Cooperates With Kras to Promote Pancreatic Cancer Through Suppression of p21-Dependent Growth Arrest. Gastroenterology 2010;139:586-97. [Crossref] [PubMed]
- Shen Z, Wen XF, Lan F, et al. The tumor suppressor gene LKB1 is associated with prognosis in human breast carcinoma. Clin Cancer Res 2002;8:2085-90. [PubMed]
- Gill RK, Yang SH, Meerzaman D, et al. Frequent homozygous deletion of the LKB1/STK11 gene in non-small cell lung cancer. Oncogene 2011;30:3784-91. [Crossref] [PubMed]
- Bonanno L, De Paoli A, Zulato E, et al. LKB1 Expression Correlates with Increased Survival in Patients with Advanced Non-Small Cell Lung Cancer Treated with Chemotherapy and Bevacizumab. Clin Cancer Res 2017;23:3316-24. [Crossref] [PubMed]
- Kitajima S, Ivanova E, Guo S, et al. Suppression of STING Associated with LKB1 Loss in KRAS-Driven Lung Cancer. Cancer Discov 2019;9:34-45. [Crossref] [PubMed]
- Skoulidis F, Goldberg ME, Greenawalt DM, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov 2018;8:822-35. [Crossref] [PubMed]
- Rizvi H, Sanchez-Vega F, La K, et al. Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J Clin Oncol 2018;36:633-41. [Crossref] [PubMed]
- Paz-Ares L, Langer CJ, Novello S, et al. LBA80 Pembrolizumab (pembro) plus platinum-based chemotherapy (chemo) for metastatic NSCLC: Tissue TMB (tTMB) and outcomes in KEYNOTE-021, 189, and 407. Ann Oncol 2019.30.
- Powell SF, Abreu DR, Langer CJ, et al. 1483PD Pembrolizumab (pembro) plus platinum-based chemotherapy (chemo) in NSCLC with brain metastases: Pooled analysis of KEYNOTE-021, 189, and 407. Ann Oncol 2019;30:v606-v607. [Crossref]
- Basik M, Aguilar-Mahecha A, Rousseau C, et al. Biopsies: Next-generation biospecimens for tailoring therapy. Nat Rev Clin Oncol 2013;10:437-50. [Crossref] [PubMed]
- von Elm E, Altman DG, Egger M, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) Statement: Guidelines for Reporting Observational Studies. PLoS Med 2007;4:e296. [Crossref] [PubMed]
- Paik PK, Veillon R, Cortot AB, et al. Phase II study of tepotinib in NSCLC patients with MET ex14 mutations. J Clin Oncol 2019;37:abstr 9005.
- Paik PK, Felip E, Veillon R, et al. Tepotinib in Non-Small-Cell Lung Cancer with MET Exon 14 Skipping Mutations. N Engl J Med 2020;383:931-43. [Crossref] [PubMed]
- Zulato E, Attili I, Pavan A, et al. Early assessment of KRAS mutation in cfDNA correlates with risk of progression and death in advanced non-small-cell lung cancer. Br J Cancer 2020;123:81-91. [Crossref] [PubMed]
- Diaz LA Jr, Bardelli A. Liquid biopsies: Genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [Crossref] [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci Transl Med 2014;6:224ra24. [Crossref] [PubMed]
- Bronkhorst AJ, Ungerer V, Holdenrieder S. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol Detect Quantif 2019;17:100087. [Crossref] [PubMed]
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985-90. [Crossref] [PubMed]
- Rolfo C, Mack PC, Scagliotti GV, et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J Thorac Oncol 2018;13:1248-68. [Crossref] [PubMed]
- Mitsudomi T, Hamajima N, Ogawa M, et al. Prognostic significance of p53 alterations in patients with non-small cell lung cancer: A meta-analysis. Clin Cancer Res 2000;6:4055-63. [PubMed]
- La Fleur L, Falk-Sörqvist E, Smeds P, et al. Mutation patterns in a population-based non-small cell lung cancer cohort and prognostic impact of concomitant mutations in KRAS and TP53 or STK11. Lung Cancer 2019;130:50-8. [Crossref] [PubMed]
- Jiao XD, Qin BD, You P, et al. The prognostic value of TP53 and its correlation with EGFR mutation in advanced non-small cell lung cancer, an analysis based on cBioPortal data base. Lung Cancer 2018;123:70-5. [Crossref] [PubMed]
- Tamiya A, Koh Y, Isa S, et al. Impact of somatic mutations on prognosis in resected non-small-cell lung cancer: The Japan Molecular Epidemiology for lung cancer study. Cancer Med 2020;9:2343-51. [Crossref] [PubMed]
- Gibert J, Clavé S, Hardy-Werbin M, et al. Concomitant genomic alterations in KRAS mutant advanced lung adenocarcinoma. Lung Cancer 2020;140:42-5. [Crossref] [PubMed]
- Guibert N, Jones G, Beeler JF, et al. Targeted sequencing of plasma cell-free DNA to predict response to PD1 inhibitors in advanced non-small cell lung cancer. Lung Cancer 2019;137:1-6. [Crossref] [PubMed]
- Facchinetti F, Bluthgen MV, Tergemina-Clain G, et al. LKB1/STK11 mutations in non-small cell lung cancer patients: Descriptive analysis and prognostic value. Lung Cancer 2017;112:62-8. [Crossref] [PubMed]
- Scagliotti GV, Parikh P, Von Pawel J, et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol 2008;26:3543-51. [Crossref] [PubMed]
- Vernieri C, Ganzinelli M, Rulli E, et al. LKB1 mutations are not associated with the efficacy of first- and second-line chemotherapy in patients with advanced non-small cell lung cancer (aNSCLC): a post hoc analysis of the TAILOR trial. ESMO Open 2020;5:e000748. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Skoulidis F, Arbour KC, Hellmann MD, et al. Association of STK11/LKB1 genomic alterations with lack of benefit from the addition of pembrolizumab to platinum doublet chemotherapy in non-squamous non-small cell lung cancer. J Clin Oncol 2019;37:abstr 102.
- Arbour KC, Jordan E, Kim HR, et al. Effects of co-occurring genomic alterations on outcomes in patients with KRAS-mutant non-small cell lung cancer. Clin Cancer Res 2018;24:334-40. [Crossref] [PubMed]
- Bange E, Marmarelis ME, Hwang WT, et al. Impact of KRAS and TP53 Co-Mutations on Outcomes After First-Line Systemic Therapy Among Patients With STK11 -Mutated Advanced Non-Small-Cell Lung Cancer. JCO Precis Oncol 2019;3:PO.18.00326.
- Yu HA, Sima CS, Shen R, et al. Prognostic Impact of KRAS Mutation Subtypes in 677 Patients with Metastatic Lung Adenocarcinomas. J Thorac Oncol 2015;10:431-7. [Crossref] [PubMed]
- Canon J, Rex K, Saiki AY, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019;575:217-23. [Crossref] [PubMed]
- Herbst RS, Schlessinger J. Small molecule combats cancer-causing KRAS protein at last. Nature 2019;575:294-5. [Crossref] [PubMed]
- Mahoney CL, Choudhury B, Davies H, et al. LKB1/KRAS mutant lung cancers constitute a genetic subset of NSCLC with increased sensitivity to MAPK and mTOR signalling inhibition. Br J Cancer 2009;100:370-5. [Crossref] [PubMed]
- Ji H, Ramsey MR, Hayes DN, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature 2007;448:807-10. [Crossref] [PubMed]
- Barber GN. STING: infection, inflammation and cancer. Nat Rev Immunol 2015;15:760-70. [Crossref] [PubMed]
- Givechian KB, Garner C, Benz S, et al. An immunogenic NSCLC microenvironment is associated with favorable survival in lung adenocarcinoma. Oncotarget 2019;10:1840-9. [Crossref] [PubMed]
- Reed JR, Stone MD, Beadnell TC, et al. Fibroblast Growth Factor Receptor 1 Activation in Mammary Tumor Cells Promotes Macrophage Recruitment in a CX3CL1-Dependent Manner. Li Y, editor. PLoS One 2012;7:e45877.
- Hato T, Zhu AX, Duda DG. Rationally combining anti-VEGF therapy with checkpoint inhibitors in hepatocellular carcinoma. Immunotherapy 2016;8:299-313. [Crossref] [PubMed]
- Ohm JE. VEGF inhibits T-cell development and may contribute to tumor-induced immune suppression. Blood 2003;101:4878-86. [Crossref] [PubMed]
- Reisländer T, Lombardi EP, Groelly FJ, et al. BRCA2 abrogation triggers innate immune responses potentiated by treatment with PARP inhibitors. Nat Commun 2019;10:3143. [Crossref] [PubMed]
- Jenzer M, Keß P, Nientiedt C, et al. The BRCA2 mutation status shapes the immune phenotype of prostate cancer. Cancer Immunol Immunother 2019;68:1621-33. [Crossref] [PubMed]
- Wen WX, Leong C-O. Association of BRCA1- and BRCA2-deficiency with mutation burden, expression of PD-L1/PD-1, immune infiltrates, and T cell-inflamed signature in breast cancer. Fei P, editor. PLoS One 2019;14:e0215381.
- Mazieres J, Drilon A, Lusque A, et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET registry. Ann Oncol 2019;30:1321-8. [Crossref] [PubMed]
- Bria E, Pilotto S, Amato E, et al. Molecular heterogeneity assessment by next-generation sequencing and response to gefitinib of EGFR mutant advanced lung adenocarcinoma. Oncotarget 2015;6:12783-95. [Crossref] [PubMed]
- Rosell R, Sureda BM, Costa C, et al. Concomitant Actionable Mutations and Overall Survival (OS) in Egfr-Mutant Non-Small-Cell Lung Cancer (NSCLC) Patients (P) Included in The Eurtac Trial: EGFR L858R, EGFR T790M, TP53 R273H and EML4-ALK (V3). Ann Oncol 2012;23:ixe22. [Crossref]
- Arbour K, Shen R, Plodkowski A, et al. MA19.09 Concurrent Mutations in STK11 and KEAP1 is Associated with Resistance to PD-(L)1 Blockade in Patients with NSCLC Despite High TMB. J Thorac Oncol 2018;13:S424. [Crossref]
- Skoulidis F, Byers LA, Diao L, et al. Co-occurring Genomic Alterations Define Major Subsets of KRAS-Mutant Lung Adenocarcinoma with Distinct Biology, Immune Profiles, and Therapeutic Vulnerabilities. Cancer Discov 2015;5:860-77. [Crossref] [PubMed]
- Padmanabhan B, Tong KI, Ohta T, et al. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell 2006;21:689-700. [Crossref] [PubMed]
- Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;21:689-700.
- Singh A, Misra V, Thimmulappa RK, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med 2006;3:e420. [Crossref] [PubMed]
- Nadal E, Palmero R, Muñoz-Pinedo C. Mutations in the Antioxidant KEAP1/NRF2 Pathway Define an Aggressive Subset of NSCLC Resistant to Conventional Treatments. J Thorac Oncol 2019;14:1881-3. [Crossref] [PubMed]
- Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018;362:eaar3593. [Crossref] [PubMed]


