
Effect of prior therapy on tumor mutational burden in NSCLC
Introduction
Higher nonsynonymous tumor mutation burden (TMB), a potential surrogate for neoantigen load, correlates with efficacy of immune checkpoint inhibitors. In patients with advanced non-small cell lung cancer (NSCLC) treated with pembrolizumab, higher TMB was associated with higher response rate and superior progression-free survival (1). Higher TMB also predicted higher response to the combination of nivolumab and ipilimumab independent of programmed death ligand (PD-L1) expression (2). The role of TMB as a potential biomarker continues to evolve, but there are significant barriers to clinical implementation. One of the major challenges is in its basic interpretation, as many different assays have been used to report TMB. These assays employ various methods and neither the mutation calls nor the cutoffs are consistent across assays (3). Another unknown variable is the impact of prior therapy on testing. Cytotoxic chemotherapy and ionizing radiation induce DNA damage. Here, we explored the impact of prior chemotherapy and/or radiation on TMB in patients with NSCLC.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-1076).
Methods
Patient selection
An institutional review board (IRB)-approved, retrospective analysis of tumor samples submitted for molecular profiling at a CLIA-certified genomics laboratory (Caris Life Sciences, Phoenix, AZ) was conducted. Tumor samples from patients with NSCLC that had undergone next generation sequencing using the Illumina NextSeq platform and TMB analysis from July, 2011 to June, 2018 were identified. Prior to molecular analysis, H&E-stained sections of formalin-fixed paraffin-embedded (FFPE) tumor tissue were manually assessed by board-certified pathologists for tumor cell populations and harvested using manual microdissection to enrich the sample to a minimum of 20% tumor nuclei.
NGS and TMB
NGS was performed on genomic DNA isolated from FFPE tumor samples using the NextSeq platform (Illumina, Inc.). A custom-designed SureSelect XT assay was used to enrich 592 whole-gene targets (Agilent Technologies). All variants were detected with >99% confidence based on allele frequency and amplicon coverage, with an average sequencing depth of coverage of 750 and an analytic sensitivity of 5%. Prior to molecular testing, tumor enrichment was achieved by harvesting targeted tissue using manual microdissection techniques. Genetic variants identified were interpreted by board-certified molecular geneticists and categorized as “pathogenic”, “presumed pathogenic”, “variant of unknown significance”, “presumed benign” or “benign” according to the American College of Medical Genetics and Genomics (ACMG) standards. When assessing mutation frequencies of individual genes, “pathogenic” and “presumed pathogenic” were counted as mutations, whereas “benign”, “presumed benign” variants, and “variants of unknown significance” were excluded. The TMB algorithm included all coding variants (missense, nonsense, frameshift, in-frame InDels) identified by NGS; exceptions were synonymous mutations and any single nucleotide polymorphisms that were described as germline in the updated reference database.
Treatment history and clinical characteristics
Through individual chart review, we determined whether patients had received any cytotoxic chemotherapy or radiation therapy in the year prior to collection of the tissue used for TMB analysis. This was based on the date of tissue sampling, not submission for molecular testing. Other key demographics, such as smoking status, were also collected.
Statistical analysis
TMB was compared between groups, adjusting for smoking status, using a likelihood ratio test of deviance. A parent regression model was fit to predict TMB values from smoking status. Extended models were then fit to predict TMB values from smoking status plus a comparison group of interest. The deviance between the nested models was compared using a likelihood ratio test. P values indicate the significance of a group’s contribution to the prediction of TMB after adjusting for smoking status.
Ethics statement
All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the MedStar Health Research Institute-Georgetown University Oncology Institutional Review Board (IRB #: 2017-0829). Because of the retrospective nature of the research, the requirement for informed consent was waived.
Results
Sample population
A total of 1,118 samples from patients with NSCLC were screened. Tumor specimens in which the same block was previously tested (n=8) were excluded. Thus, each sample corresponded to an individual patient. In addition, samples in which treatment history (n=86) and/or TMB data (n=45) were not available were excluded. One sample was consistent with adenoid cystic carcinoma upon histology review and was not included in the analysis. We also adjusted for smoking status which was not available for n=8 cases. The final TMB analysis was performed on 970 samples (Figure 1).
Patient and tumor characteristics
The majority of the samples (n=815) were collected from patients that had not received any prior therapy (treatment-naïve). A total of 155 samples were collected from patients who were previously treated with chemotherapy and/or radiation (treatment-experienced). 128 patients received chemotherapy and 85 patients received radiation. Table 1 describes the patient and tumor characteristics. The median age was statistically higher in the treatment-naïve cohort (67 years; range, 25–93) compared to the treatment-experienced cohort (63 years; range, 24–87) (P<0.001). Treatment-naïve samples were more likely to be acquired from the primary tumor than a distant metastasis. In contrast, 67% of the treatment-experienced samples were acquired from a metastatic site (P<0.001). In both treatment groups, approximately 50% of patients were former smokers. There was a higher percentage of lifelong never smokers in the treatment-experienced group (21%) compared to those having received no prior treatment (13%). Histology was balanced between the treatment cohorts, with the most common histology being adenocarcinoma in both groups.
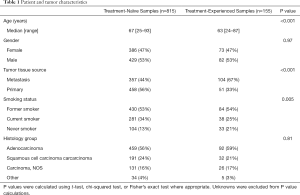
Full table
TMB comparison across cohorts
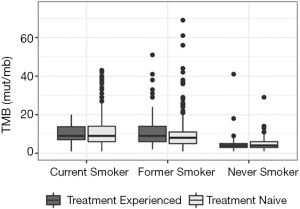
TMB was calculated on 970 tumor specimens. Median TMB was statistically higher in individuals younger than 65 compared to those older than 65 (8 vs. 7 mut/Mb, P=0.01), and in males compared to females (8 vs. 7 mut/Mb, P=0.02). As expected, current and former smokers had statistically higher TMB than never smokers (median 9 vs. 9 vs. 4 mut/Mb, P<0.001).
The median TMB was 8 mut/Mb in both treatment-naïve and treatment-experienced samples (Figure 2). Additional analysis of the treatment-naïve versus treatment-experienced groups by smoking status revealed no significant difference in TMB among never smokers and current smokers (P=0.55 and P=0.87; adjusted for multiple testing P=0.68 and P=0.93, respectively). However, former smokers had a slightly higher TMB in the pre-treated cohort (P=0.025; adjusted for multiple testing P=0.037). Further evaluation of TMB by chemotherapy treatment status showed a median TMB of 8 mut/mb (95% CI: 7.5–10.0) in chemotherapy-naïve and 8.5 mut/mb (95% CI: 8.0–8.5) in the chemotherapy-experienced samples. After adjusting for smoking, there was no statistically significant difference in TMB between the chemotherapy-naïve and chemotherapy-experienced samples (P=0.262). The median TMB was 8 mut/mb (95% CI: 8.0–8.5) in radiation-naïve samples and 9 mut/mb (95% CI: 8.0–11.0) in radiation-experienced samples, which was also not significantly different (overall adjusted for smoking P=0.129) (Table 2). TMB was slightly higher within patients with prior radiation within the former smoker cohort, but was not statistically significant across any smoking status.

Full table
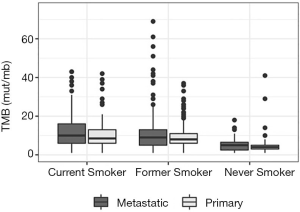
TMB was significantly higher when specimen source was from a metastatic site with a median TMB of 8 mut/mb (95% CI: 8.5–9.5) compared to the primary tumor site at 7 mut/Mb (95% CI: 7.5–8.5) (adjusted for smoking P=0.007) (Figure 3). However, when comparing TMB between the treatment-naïve and treatment experienced cohorts within samples from the same specimen source, there was no statistically significant difference (P=0.842) (Table 3).

Full table
TMB was lower in oncogene-driven NSCLC compared to wild-type NSCLC (median TMB of 4 mut/mb (95% CI: 4.4–5.5) vs. 9 mut/mb (95% CI: 8.5–9.4) (P<0.001). Genetic alterations included in the analysis were EGFR or BRAF mutations, and fusions in ALK, ROS1, RET, or NTRK. TMB was also compared between the treatment-naïve and treatment experienced cohorts after adjusting for the presence on an oncogene. There was no significant difference in TMB between treatment cohorts after accounting for oncogene status (P=0.248) (Table 4). Within the oncogene mutated subgroup, the median TMB was 4 mut/mb (95% CI: 4.0–6.0) in never smokers, 4 mut/mb (95% CI: 3.5–4.5) in former smokers, and 6.5 mut/mb (95% CI: 6.0–11.0) in current smokers. In the oncogene wild-type subgroup, the median TMB was 9 mut/mb for both the treatment-naïve (95% CI: 8.5–9.5) and treatment-experienced groups (95% CI: 9.0–11.5) (P=0.124). Within the oncogene positive patients, median TMB was 4 mut/mb (95% CI: 4.0–6.0) and 4.5 mut/mb (95% CI: 4.5–5.5) for pre-treated and treatment-naïve cohorts (P=0.836).

Full table
Discussion
TMB is an evolving biomarker for response to immunotherapy. Somatic mutational and neoantigen burden is associated with superior outcomes with immune checkpoint inhibitors (1). In multiple retrospective and prospective efforts, higher TMB has been associated with higher response rates and longer PFS in patients with NSCLC receiving checkpoint inhibitors. The role of TMB as a predictive factor for survival, however, is unclear. One of the challenges of biomarker development is controlling for all relevant clinical variables. Here, we demonstrate that prior DNA damaging therapy is not a relevant variable. A history of prior cytotoxic chemotherapy or prior radiation therapy was not associated with a difference in measured TMB. This is in part because chemotherapy and radiation induce subclonal mutations, those present only in a subset of the tumor, as opposed to clonal mutations (4-7).
In contrast, TMB was higher when the sample was acquired from a metastatic site compared to the primary tumor. This is consistent with prior reports that TMB surpasses the threshold of 10 mut/Mb more frequently in metastatic sites (38%) as opposed to primary sites (25%) in lung adenocarcinoma. Furthermore, TMB also varies by the site of metastasis; brain metastasis had the highest likelihood of exceeding 10 mut/mb (61% vs. 35% for other metastasis, P<0.001) (8). In another matched analysis, TMB from metastatic tissue samples was 36% higher than from primary tissue samples (paired Wilcoxon, P=0.0008). However, the predictive value of TMB in terms of overall survival was equivalent regardless of tissue source (9). Together with these other studies, our data supports the consideration of tissue source in interpreting TMB values.
As expected, TMB was lower in oncogene-driver positive NSCLC compared to wild-type NSCLC (10). The lack of variation in TMB in patients treated with chemotherapy and/or radiation was consistent after adjusting for the presence of an oncogene. Although TMB does not seem to play a predictive role in this manner, other data suggests that higher TMB is negatively correlated with clinical outcomes in metastatic EGFR-mutant lung cancer treated with EGFR-TKI (11). The implications of TMB in oncogene-driven lung cancer needs further clarification.
As a retrospective analysis, our study has several limitations. Although the analysis was adjusted for smoking status, the arms were not matched. It would be important to distinguish whether any change in TMB would be detectable in the same individual before and after chemotherapy and/or radiation. In another study, a paired analysis of pre and post-chemotherapy tumor tissue specimens was completed in 13 patients. Consistent with the results of our study, there was no significant different difference in TMB (12). In addition, the exact chemotherapy agent or type of radiation was not differentiated in our dataset. Whether specific classes of chemotherapy or aspects of radiation therapies would impact TMB is unknown. This study did not assess the impact of DNA damaging agents more than 1 year from tumor sampling which may be relevant in radiation or chemotherapy induced cancers.
To summarize, prior exposure to chemotherapy and/or radiation was not associated with a significant difference in TMB even after accounting for all coding variants in TMB analysis. This result was also true after adjusting for the presence of an oncogene. As previously documented, TMB was higher in tissue from metastatic tumor sites compared to primary sites. Future analyses may need to control for the site of sample acquisition, though prior therapy does not appear relevant.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-1076
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-1076
Peer Review File: Available at http://dx.doi.org/10.21037/tlcr-20-1076
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-1076). SVL serves as an unpaid editorial board member of Translational Lung Cancer Research from Jan 2020-Dec 2021. AV reports personal fees from Caris Life Sciences, outside the submitted work. JN reports grants from Merck, personal fees from Astra Zeneca, personal fees from Genentech, personal fees from Western Oncolytics, outside the submitted work. KP reports personal fees from Caris Life Sciences, during the conduct of the study; personal fees from Caris Life Sciences, outside the submitted work; and Full Time Employee of Caris Life Sciences. MS reports other from Caris Life Sciences, during the conduct of the study; other from Caris Life Sciences, outside the submitted work. SVL reports grants from Alkermes, grants, personal fees and non-financial support from AstraZeneca, grants from Bayer, grants and personal fees from Blueprint, non-financial support from Boehringer-Ingelheim, grants and personal fees from Bristol-Myers Squibb, personal fees from Catalyst, personal fees from Celgene, grants from Clovis, grants from Corvus, personal fees from G1 Therapeutics, grants, personal fees and non-financial support from Genentech/Roche, personal fees from Guardant Health, personal fees from Inivata, grants and personal fees from Janssen, grants and personal fees from Lilly, grants from Lycera, personal fees from LOXO, grants, personal fees and non-financial support from Merck/MSD, grants from Merus, grants from Molecular Partners, grants and personal fees from Pfizer, personal fees from PharmaMar, grants from Rain Therapeutics, personal fees from Regeneron, grants from RAPT, personal fees from Spectrum, grants from Takeda, grants from Turning Point Therapeutics, outside the submitted work. The other have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the MedStar Health Research Institute-Georgetown University Oncology Institutional Review Board (IRB #: 2017-0829). Because of the retrospective nature of the research, the requirement for informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Hellmann MD, Ciuleanu T-E, Pluzanski A, et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med 2018;378:2093-104. [Crossref] [PubMed]
- Fancello L, Gandini S, Pelicci PG, et al. Tumor mutational burden quantification from targeted gene panels: major advancements and challenges. J Immunother Cancer 2019;7:183. [Crossref] [PubMed]
- Pich O, Muiños F, Lolkema MP, et al. The mutational footprints of cancer therapies. Nat Genet 2019;51:1732-40. [Crossref] [PubMed]
- Johnson BE, Mazor T, Hong C, et al. Mutational Analysis Reveals the Origin and Therapy-Driven Evolution of Recurrent Glioma. Science 2014;343:189-93. [Crossref] [PubMed]
- Murugaesu N, Wilson GA, Birkbak NJ, et al. Tracking the Genomic Evolution of Esophageal Adenocarcinoma through Neoadjuvant Chemotherapy. Cancer Discov 2015;5:821-31. [Crossref] [PubMed]
- McGranahan N, Furness AJS, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016;351:1463-9. [Crossref] [PubMed]
- Stein MK, Pandey M, Xiu J, et al. Tumor Mutational Burden Is Site Specific in Non–Small-Cell Lung Cancer and Is Highest in Lung Adenocarcinoma Brain Metastases. JCO Precis Oncol 2019;1-13. [Crossref]
- Schnidrig D, Turajlic S, Litchfield K. Tumour mutational burden: primary versus metastatic tissue creates systematic bias. Immuno Oncol Technol 2019;4:8-14. [Crossref]
- Rizvi H, Sanchez-Vega F, La K, et al. Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J Clin Oncol 2018;36:633-41. [Crossref] [PubMed]
- Offin M, Rizvi H, Tenet M, et al. Tumor Mutation Burden and Efficacy of EGFR-Tyrosine Kinase Inhibitors in Patients with EGFR-Mutant Lung Cancers. Clin Cancer Res 2019;25:1063-9. [Crossref] [PubMed]
- Sakai H, Takeda M, Sakai K, et al. Impact of cytotoxic chemotherapy on PD-L1 expression in patients with non–small cell lung cancer negative for EGFR mutation and ALK fusion. Lung Cancer 2019;127:59-65. [Crossref] [PubMed]



