
HER2 transmembrane domain mutation: comprehensive characteristics and real-world evidence of treatment response in Chinese lung adenocarcinoma
Introduction
The comprehensive management of non-small cell lung cancer (NSCLC) has been revolutionized, switching from a one-treatment-fits-all approach to personalized medicine (1). With the advancements in next-generation sequencing (NGS), numerous driver mutations have been revealed and treatments have attained higher precision (2). Tumor NGS reports are detailed into specific alterations and mutation loci, as mutations at different loci of the same gene might lead to different treatment responses (3). Mutations at rare loci acting as driver mutations have also been found.
Erb-b2 receptor tyrosine kinase 2 (HER2/ERBB2) is one of the driver genes identified that are susceptible to targeted treatments (4). HER2 alteration is usually seen as amplification (2–23%) and/or overexpression (11–32%) in lung adenocarcinoma (LUAD), but rarely as mutations (1.6–4%) (5-7). HER2 mutation is a distinct therapeutic target and a poor prognosis predictor (8-10). Most of the HER2-mutant NSCLCs are adenocarcinomas and occur in female never-smokers (8,11,12). Overall survival of HER2-mutant NSCLCs is 28.4 months (8) and ranges from 19.2 to 22.8 months among those diagnosed at stage IV (11,13). The gene product of HER2 is a membrane protein by the same name, which is a receptor tyrosine kinase that forms homodimers or heterodimers to activate downstream pathways (14) and has a transmembrane domain (TMD) where intramolecular interactions occur and lead to dimerization and activation (14). In LUAD, most HER2 mutations occur in the kinase domain (KD, 2–4%), but rarely in the TMD (0.07–0.20%) (8,15-19). Moreover, activating TMD mutations most frequently affect residues V659 and G660 in exon 17 (20-23).
Most HER2-targeting agents such as trastuzmab did not result in comparable efficacy in NSCLC harboring HER2 exon 20 mutations as they did in HER2-positive breast cancer. A mechanistic explanation is yet to be established but may implicate intratumoral heterogeneity and lower HER2 expression in HER2-mutant NSCLC compared with in HER2-amplified breast cancer (24). Theraies for HER2-mutant NSCLC is under active development, among which the most promising include reversible or irreversible anti-HER2 TKIs and antibody-drug conjugates (ADCs) that target HER2 alone or along with EGFR (25). Patients harboring a HER2 TMD mutations were reported to respond to HER2 tyrosine kinase inhibitor (TKI), lapatinib, and afatinib in case reports (15,26); and to the antibody-drug conjugate, ado-trastuzumab emtansine (TDM-1) in a basket trial (9). Due to their low prevalence, it remains to be determined whether TMD mutations would result in a comparable response to targeted therapy as HER2 KD mutations.
Herein, we demonstrated the comprehensive characteristics and prognosis of the HER2 TMD-mutant population and illustrated the real-world evidence (RWE) of treatment response from the largest reported LUAD cohort that harbors HER2 V659E mutation. To enrich the limited clinical data on how to overcome resistance to targeted therapy, we postulated to challenge the resistance of former-line TKI with TKI that has stronger binding energy to the ATP-pocket of HER2 KD mutants. Additionally, we demonstrated a three-month response to the off-label use of pyrotinib in fifth-line therapy. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tlcr-21-107).
Methods
Database, sample collection, and NGS
We screened the genomic profiles of samples from 7,812 Chinese LUAD patients in the Burning Rock LAVA Open-access Database, profiled between Jan 2014 and July 2019. As an external cohort, a total of 4,587 samples of 4,185 patients was selected from The Cancer Genome Atlas (TCGA) and other studies (27-37) through the open platform cBioPortal for Cancer Genomics (38,39). After deduplication, a total of 2,966 patients with 3,480 samples were included. In brief, 38 HER2-mutant patients with overall survival (OS) were deduplicated and included in the final analysis. Protocols of sample collection, DNA extraction, and plasma cell-free DNA preparation were as previously described (40). Captured-based targeted NGS using various panels, including panels with eight, 168, or 520 cancer-related genes (Burning Rock Biotech, Guangzhou, China) were performed according to protocols as previously described (41). The panel gene lists are shown in Tables S1-S3. TMD is defined as 27 amino acids (Ala648 to Leu674), and KD is defined as 274 amino acids (Ile714 to Val987) (42).
Clinical characteristics and response assessment
Patient characteristics, treatments, and outcomes were obtained from the LAVA database with the permission of patients and their physicians in-charge. Patients were followed from the date of diagnosis until death or the last available follow-up. Response to therapy was measured using RECIST v1.1 criteria and OS was calculated from the date of the initiation of treatment until the day of last follow-up or death. All procedures performed in studies involving human participants were in accordance with the Helsinki Declaration (as revised in 2013). The study was conducted under the approval of the Ethics Committee of Peking Union Medical College Hospital (ZS-1329). Written informed consent was obtained from each patient.
Computational methods of simulation and energy analysis of lapatinib, afatinib, and pyrotinib in HER2 KD binding
AutoDock 4.2 (43) was used for molecular docking simulations of lapatinib, afatinib, and pyrotinib and predicting binding affinity with the HER2 KD (PDB code:3RCD). The Lamarckian genetic algorithm (LGA) (44) was used to optimize the binding conformations of compounds. The structure of the lowest predicted binding free energy in the most popular cluster of each compound was selected as the initial conformation for the following molecular dynamic (MD) simulation. MD simulations were implemented by AMBER18 (45). The potentials for protein and ligands in each complex were generated based on AMBER03 (parm03) forcefield (46), and general amber forcefield (gaff) (47), respectively. Partial atomic charges were assigned using the AM1-BCC charge method (48,49) within the ANTECHAMBER in AMBERTools12 (50). The compound was neutralized with the counterions of Cl− and the whole system was immersed in a truncated octahedron’s box of TIP3PBOX water (51). The periodic boundary is 12 A from any solute atoms. The binding free energy was evaluated by the MMGBSA method, which is based on the following equation (52):
ΔGbind = Gcomplex–Gprotein–Gligand = ΔEMM+ΔGGB+ΔGSA–TΔS = ΔEvdw+ΔEele+ΔGGB+ΔGSA–TΔS
where ΔEMM is the molecular mechanics interaction energy between the protein and the ligand, which is comprised of two parts: the electrostatic (ΔEele) and the van der Waals energies (ΔEvdW). Further, ΔGGB and ΔGSA are the polar and nonpolar contributions of the desolvation free energy upon the construction of the protein-substrate complex. The 200 snapshots taken from the last 6.0 ns MD simulation trajectories of the complex were used to calculate the protein-substrate binding free energy, which was accomplished by the MMPBSA.py program in AmberTools18 (53).
Statistical analysis
Clinical characteristics between the group of HER2 TMD-mutant patients with OS data and the group of HER2 non-TMD mutation with OS data were performed with Student t-test if the characteristic was a continuous variable; with Chi-square test if the characteristic was a binary variable; and with non-parametric Mann-Whitney test if the characteristic was a ranked variable. Kaplan Meier curves for OS of the two above-mentioned groups were plotted. Pairwise comparisons using log-rank test were performed and the P value was adjusted using the Benjamini-Hochberg method. Cox regression model was used in the univariate and multivariate analysis. All the above-mentioned statistical analyses were conducted in R Studio using Package survminer (v0.4.7; http://cran.r-project.org/web/packages/survminer/index.html), ggplot2 (54), and survival (55).
Results
Comprehensive characteristics of the HER2 TMD mutated LUAD
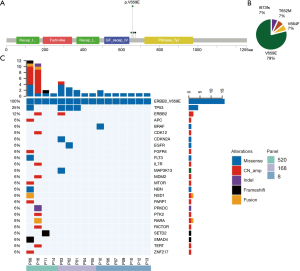
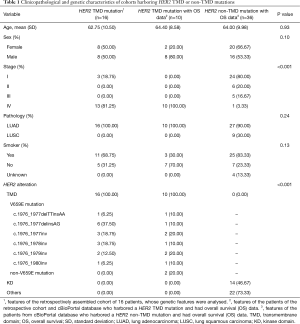
We analyzed the NGS data of 7,812 LUAD patients and identified 14 (0.18%) with a HER2 TMD mutation. These TMD mutations covered four amino acids among the 27 amino acids of the HER2 TM domain (Figure 1A,B). The most mutations occurred at codon 659, which were mostly V659E mutation (11/7,812, 0.14%). Seven patients from the external cohort of 2,966 NSCLC patients harbored a HER2 TMD mutation (0.24%) and three of these harbored a HER2 V659E mutation (0.10%). A retrospective search in medical records from cooperating medical centers assembled a cohort of 16 HER2 V659E-mutant patients (Table 1), and detailed characteristics of the patients are shown in Table S4. Five of these patients were not included in the formal analysis, and eight did not have follow-up information. In this cohort, the tumors of all patients were LUAD and harbored a dinucleotide missense mutation, including 10 TT→AA and 6 TT→AG (c.1976_1977). There were equal numbers of males and females (n=8 for each), and 68.75% were non-smokers (n=11). Most of the tumors harboring HER2 V659E were diagnosed at an advanced stage (stage III–IV, n=13/16, 81.25%). Among the 16 concomitant genetic alterations (Figure 1C), TP53 was the most frequently mutated gene (n=4, 25.0%) followed by HER2 amplification (n=2, 12.5%), NSD1 (n=2, 12.5%) and RARA (n=2, 12.5%). The sequencing results of patient 02 and 10 suggested that HER2 V659E could coexist with the driver mutations EGFR (p.T783I) and BRAF (p.I659M), though not at their hotspots. The eight patients from the in-house cohort with OS information together with two patients from the external cohort harboring HER2 TMD mutations and with available OS were assembled into a cohort for further survival analysis. Thirty-six patients from the external cohort harboring HER2 non-TMD mutation were also analyzed as a comparative cohort. These two cohorts were significantly different in the stage of diagnosis (P<0.001) but were comparable from other perspectives.
Full table
Real-world evidence of treatments for HER2 V659E patients and responses
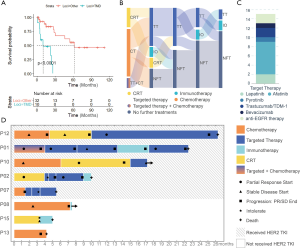
Follow-up treatment information was available for eight HER2 V659E-mutant patients from the in-house cohort, and OS data were available for all 46 patients harboring HER2 mutations from both in-house and external cohorts. The Kaplan-Meier curves of patients from HER2 TMD mutation and non-TMD mutation cohorts are shown in Figure 2A. The median OS was 10.0 months (95% CI, 5.0–NA) for HER2 TMD-mutant patients, and 61.6 months (95% CI, 30.1–NA) for patients harboring non-TMD HER2 mutations. Table 2 presents the hazard ratios (HR) associated with clinical and genetic characteristics. OS was significantly worse for HER2 TMD-mutant patients than non-TMD-mutant patients (HR =7.9, 95% CI: 1.0–61.0, P=0.046). Stage was associated with survival but was not statistically significant in Cox multivariate survival analysis (P=0.47).

Full table
Most of the HER2 TMD-mutant patients (7/8, 87.50%) received more than one line of treatment (Figure 2B) and all patients received chemotherapy or combination therapy including chemotherapy as their first-line treatment. Among treatment lines that used only chemotherapy or concurrent chemoradiotherapy, only two lines of treatment in two patients achieved partial response (ORR 22.2%, 2/9). Five patients received targeted therapy as at least one line of therapy and four of these achieved longer OS than the others. However, the choice of targeted therapy was not quite standard. Two patients received EGFR-TKI gefitinib, which was not guideline-recommended, and did not respond (Figure 2C). One patient received anti-angiogenesis agent bevacizumab, albeit for only one month before disease progession (Figure 2C). The overall response rate (ORR) of targeted therapy in assessable treatment lines was 57.1% (4/7). The longest progression-free survival (PFS) was 16 months, which was achieved by targeted therapy (afatinib) in P12 (Figure 2D). However, afatinib was also the most intolerable treatment, and two patients stopped using it because of severe oral ulcers, diarrhea, vomiting, or rash. Four patients received immunotherapy alone as a line of therapy and none of these achieved partial relief during or after the treatment (ORR 0.0%).
A case report of a patient who is responding to pyrotinib as a fifth-line therapy
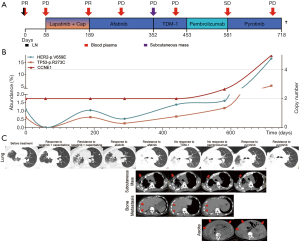
Patient 01, a 50-year-old Chinese woman with no smoking history, was diagnosed with stage IVa (pT2bN3M1a) LUAD with a rare HER2 V659E mutation (Figure 3). PET-CT revealed a 4.5 cm × 5.3 cm × 3.4 cm mass in the right lower lobe, small, scattered nodules in the left lung, lymph node metastases, and a large amount of malignant pleural effusion. She was treated with lapatinib (1,250 mg, once daily) and capecitabine (2,000 mg, divided into twice daily) as first-line therapy, achieving a partial response (PR) with a PFS of 3 months, after which she presented with fast progression in her primary tumor and chest wall metastasis. Subsequently, afatinib (40 mg daily) was administered as second-line therapy and achieved PR with a PFS of 5 months. At progression, her bone and subcutaneous metastases developed, and subcutaneous tumor tissue samples were sent for NGS testing revealing novel amplifications in both HER2 and CCNE1. Her treatment was then switched to TDM-1 (150 mg daily) and her pain was quickly relieved, but she rapidly progressed to atelectasis and hemoptysis and was subsequently administered with pembrolizumab (90 mg every 3 weeks). While her hemoptysis and pain were significantly alleviated in a week, despite a low tumor mutation burden (TMB), CT showed inflammation and no improvement in the primary nor the metastasized nodules in the lungs and new lesions were seen in the liver, adrenal gland, and abdominal wall. She then received a novel dual EGFR/HER2 inhibitor, pyrotinib, achieving fast reduction of ascites and pain relief and her primary tumor and metastatic tumors in the lungs and adrenal gland shrank. Metastatic lesions of the chest wall, bones, subcutaneous tissue, liver, and abdominal wall were also stable. After 3 months of PR, a fast progression in the lung was observed accompanied by the development of ascites and a significant increase in circulating tumor DNA (ctDNA) mutation abundance. The patient passed away shortly after.
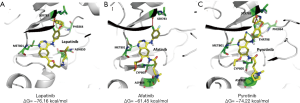
Structure and energy analysis of lapatinib, afatinib, and pyrotinib in HER2 kinase domain binding
Lapatinib is a non-covalent inhibitor that interacts with HER2 dynamically as Eq. [1], and afatinib and pyrotinib are covalent inhibitors whose HER2-interaction follows Eq. [2] (E = target, I = inhibitor, [EI] = non-covalent binding state of the target and the inhibitor, EI* = covalently bonded state of the target and the inhibitor). Thus, afatinib and pyrotinib are more favorable for blocking HER2 KD from being continuously phosphorylated than lapatinib. Between the two covalent inhibitors, pyrotinib (ΔGPyrotinib = −65.92 kcal/mol) shows stronger non-covalent binding ability than afatinib (ΔGAfatinib = −59.71 kcal/mol). ΔG calculations of the three TKIs are shown in Table S5 and docking simulation is shown in Figure 4A,B,C.
Discussion
In this study, the incidence of HER2 TMD mutation was 0.18% (14/7,812) in the Chinese LUAD population. These TMD mutations were more frequent in NSCLC than other solid tumors, and with a higher prevalence in LUAD than lung squamous cell carcinoma (LUSC) (42,56). V659E was the most recurrent mutation compared to that found in colorectal cancer, which was I655V (57). The prevalence of V659E was 0.14% (11/7,812), higher than the mutation rate in the external cohort derived from the TCGA database and ten other studies through cBioPortal (0.10%, 3/2,966) (27-37). The prevalence was also higher than 0.009%, which was reported by Ou et al. in 2017 (15). This variation might be explained by differences in the size or race of the investigated cohort. In addition to our current knowledge of all HER2 KD mutations in published studies that occurred in LUAD (8), all TMD mutations identified in our study also were detected in LUAD and all the HER2 V659E mutations in our cohort resulted from a dinucleotide missense mutation. It was reported that HER2 mutation occurred more frequently in younger women and non-smokers (6,8,11,58-60), and rarely co-existed with other lung cancer driver mutations (61). However, we observed no significant difference in gender or smoking history. We also found two patients harboring both HER2 mutation and EGFR or BRAF mutations, respectively. This accorded with a previous study that non-KD mutations did not exclude concurrent driver mutations as KD mutations did (8).
Previous studies had reported a shorter OS in HER2-mutant lung cancer, especially KD mutations (8,12). The present study found that patients harboring TMD mutations had even shorter OS compared to HER2 non-TMD mutations with statistical significance (HR 7.9, 95% CI, 1.03–61, P<0.001), including KD mutations. It was also observed that patients harboring TMD mutations were diagnosed at a more advanced stage (HR 1.2, 95% CI, 0.61–2.5, P<0.001), but were not significantly associated with a poorer prognosis.
In the present real-world retrospective cohort, although all patients had the same somatic gene mutation and similar clinical characteristics, their treatment regimens were diverse. For targeted therapy in HER2 exon 20 insertion, in vitro cell-line study demonstrated that the most common YVMA insertion is sensitive to only neratinib, poziotinib, and pyrotinib (24,25). In the clinical setting, targeted therapy is the National Comprehensive Cancer Network (NCCN) guideline-recommended treatment for HER2 mutation (4). The NCCN recommendation has evolved over time with trastuzumab, lapatinib, and pertuzumab being recommended in 2013; trastuzumab and afatinib in 2014; and TDM-1 in 2018. However, the works cited to support these recommendations did not include patients harboring HER2 TMD mutation until Li et al. published the results of their basket trial of HER2-mutant lung cancers (9). In that study, two V659E patients were recruited in the 18-patient cohort and only one of them achieved PR, while the disease progressed in the other (ORR 50%). In the present cohort, most of the patients received targeted therapies (ORR 57.1%), among which afatinib achieved the longest PFS of 16 months. However, two patients showed intolerance to afatinib and needed an appropriate substitution. Among patients who received targeted therapy, two patients progressed after receiving gefitinib. This suggested HER2 V659E mutation did not respond to gefitinib, as was demonstrated previously in vitro (62). Three patients received immunotherapy, none of whom responded. Although it was previously reported that the response rate to anti-PD-1 immunotherapy in HER2-altered NSCLC was 7–35% (63-65), and was similarly low in other driver mutations, a larger cohort is needed to accurately determine the response rate of immunotherapy in HER2 TMD-mutant patients. Chemotherapy and concurrent chemoradiotherapy were used in all patients (ORR 22.2%), and the efficacy was seen to be better than immunotherapy, although worse than targeted therapy.
We suggest the use of targeted therapy to rechallenge targeted therapy resistance. Previous studies reported short PFS in HER2 TMD-mutant patients who responded to targeted therapies. The median PFS was 5 months in a basket trial treating HER2-mutant NSCLC patients with TDM-1 (9) and PFS was reported to be in the range of 3–18 months (median 5 months) in case reports of patients with HER2 TMD-mutant patients treated with TKI (15,26,42). As we reported, a stage IV patient with multiple distal metastases achieved an OS of 22.4 months and obtained PR when receiving with first-line lapatinib with capecitabine, second-line afatinib, and fifth-line pyrotinib treatment. Each line of treatment had a PFS longer than 3 months. This might be a solution to conquer the short PFS of targeted therapy. We postulated that pyrotinib can conquer resistance to afatinib, and afatinib to lapatinib because of increased TKI binding affinity. Molecular dynamics simulation and binding free energy analysis were conducted to investigate the binding mode and strength of lapatinib, afatinib, and pyrotinib for the HER2 kinase domain to further explain the sequential conquer of drug resistance in the reported case. The irreversible binding of covalent inhibitors to the target reduces the competition of the endogenous substrate ATP on the kinase, which in turn provides more sustained kinase inhibition than non-covalent inhibitors (66,67). Thus, we conclude that the affinity of lapatinib, afatinib, and pyrotinib increases sequentially.
The efficacy of pyrotinib for tumors harboring an activating HER2 alteration is being studied in a phase I basket trial including NSCLC (68). Another study reported that LUADs harboring HER2 KD mutations could respond to pyrotinib better than afatinib (69). The results of our study suggest pyrotinib is also effective for advanced stage HER2 V659E-mutant LUAD. More refined stratification of patients based on the type of HER2 activating alterations might be needed in further studies of pyrotinib.
A limitation of this study is that it was a descriptive study based on a real-world multicenter retrospective cohort. The efficacy of targeted therapy, chemotherapy, and immunotherapy should not be directly compared. This limitation reflects the real-world challenge in assembling cohorts of HER2 TMD-mutant patients because of the low prevalence. While basket trials might serve as a solution, the intra-cohort heterogeneity of prognosis might reduce its reliability in specific mutation subgroups. We suggest finer stratification and transparent data sharing policy in further studies to facilitate future meta-analysis of individual participant data, which is considered as the most accessible high-level evidence for low prevalence mutations.
Conclusions
The present study was the first to report the comprehensive profiles and real-world evidence of treatment responses in HER2 TMD-mutant lung cancer patients and to report a case that rechallenged TKI resistance with stronger-affinity TKI and responsed to fifth-line pyrotinib.
With the wide application of NGS in tumor evaluation and the inclusion of HER2 in most NGS panels, more patients with HER2 TMD mutations have been uncovered. As we have illustrated, patients harboring HER2 TMD mutations have poorer survival than patients with other HER2 mutations, highlighting the need for finer stratification of HER2 mutations in future clinical studies.
Acknowledgments
We thank Ms. Huihui Xu from Nanopep Biotech Co., Beijing, China for her help in organizing materials.
Funding: This work was supported by the National Key Research and Development Program of China (No. 2016YFC0901500), Beijing Health Promotion Association (BJHPA-FW-XHKT-2020040400344), CAMS Innovation Fund for Medical Sciences (2020-I2M-C&T-A-003), Chinese Society of Clinical Oncology CSCO-MSD (Y-MSD2020-0270), Ministry of Science and Technology of the People’s Republic of China, Special Data Service for Oncology: The National Population and Health Scientific Data Sharing Platform (NCMI-ABD02-201809 and NCMI-YF02N-201906), and Wu Jieping Medical Foundation Precision Treatment for Thoracic and Abdominal Cancer Fund (320.6750.19092-43).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-21-107
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-21-107
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-21-107). Dr. JY, Dr. BL, Dr. HHZ, and Dr. JZ report personal fees from Burning Rock Biotechnology Co. Ltd. during the conduct of the study. Dr. NXL reports grants from National Key Research and Development Program of China, Beijing Health Promotion Association, grants from Beijing Natural Science Foundation, Major projects of the Beijing Municipal Science and Technology Commission, and Beijing Dongcheng District Science and Technology Commission Excellent Talents Project, during the conduct of the study. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the Helsinki Declaration (as revised in 2013). The study was conducted under the approval of the Ethics Committee of Peking Union Medical College Hospital (ZS-1329). Written informed consent was obtained from each patient.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Reck M, Rabe KF. Precision Diagnosis and Treatment for Advanced Non–Small-Cell Lung Cancer. N Engl J Med 2017;377:849-61. [Crossref] [PubMed]
- Nangalia J, Campbell PJ. Genome Sequencing during a Patient’s Journey through Cancer. N Engl J Med 2019;381:2145-56. [Crossref] [PubMed]
- Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR Mutation and Resistance of Non–Small-Cell Lung Cancer to Gefitinib. N Engl J Med 2005;352:786-92. [Crossref] [PubMed]
- NCCN. Non-Small Cell Lung Cancer (Version 1.2021). National Comprehensive Cancer Network. Available online: https://www.nccn.org/professionals/physician_gls/ pdf/nscl.pdf. Accessed 15 Dec 2020.
- Stephens P, Hunter C, Bignell G, et al. Intragenic ERBB2 kinase mutations in tumours. Nature 2004;431:525-6. [Crossref] [PubMed]
- Shigematsu H, Takahashi T, Nomura M, et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res 2005;65:1642-6. [Crossref] [PubMed]
- Pellegrini C, Falleni M, Marchetti A, et al. HER- 2/Neu alterations in non-small cell lung cancer: a comprehensive evaluation by real time reverse transcription-PCR, fluorescence in situ hybridization, and immunohistochemistry. Clin Cancer Res 2003;9:3645-52. [PubMed]
- Wei XW, Gao X, Zhang XC, et al. Mutational landscape and characteristics of ERBB2 in non-small cell lung cancer. Thoracic Cancer 2020;11:1512-21. [Crossref] [PubMed]
- Li BT, Shen R, Buonocore D, et al. Ado-Trastuzumab Emtansine for Patients With HER2-Mutant Lung Cancers: Results From a Phase II Basket Trial. J Clin Oncol 2018;36:2532-7. [Crossref] [PubMed]
- Pillai RN, Behera M, Berry LD, et al. HER2 mutations in lung adenocarcinomas: A report from the Lung Cancer Mutation Consortium. Cancer 2017;123:4099-105. [Crossref] [PubMed]
- Mazieres J, Peters S, Lepage B, et al. Lung cancer that harbors an HER2 mutation: epidemiologic characteristics and therapeutic perspectives. J Clin Oncol 2013;31:1997-2003. [Crossref] [PubMed]
- Tomizawa K, Suda K, Onozato R, et al. Prognostic and predictive implications of HER2/ERBB2/neu gene mutations in lung cancers. Lung Cancer 2011;74:139-44. [Crossref] [PubMed]
- Kris MG, Johnson BE, Berry LD, et al. Using Multiplexed Assays of Oncogenic Drivers in Lung Cancers to Select Targeted Drugs. JAMA 2014;311:1998. [Crossref] [PubMed]
- Mineev KS, Bocharov EV, Pustovalova YE, et al. Spatial structure of the transmembrane domain heterodimer of ErbB1 and ErbB2 receptor tyrosine kinases. J Mol Biol 2010;400:231-43. [Crossref] [PubMed]
- Ou SI, Schrock AB, Bocharov EV, et al. HER2 Transmembrane Domain (TMD) Mutations (V659/G660) That Stabilize Homo- and Heterodimerization Are Rare Oncogenic Drivers in Lung Adenocarcinoma That Respond to Afatinib. J Thorac Oncol 2017;12:446-57. [Crossref] [PubMed]
- Ma F, Zhang M, Ouyang Q, et al. ERBB2 mutation profiling with next-generation sequencing (NGS) in solid tumors. American Society of Clinical Oncology; 2018.
- Yu R, Yang SW, Bao H, et al. P2.13-46 Comprehensive Investigation of ERBB2 Transmembrane Domain Mutations (V659/G660) in 12,833 Chinese Lung Cancer Patients. Journal of Thoracic Oncology 2018;13:S816-S7. [Crossref]
- Xie F, Zhang Y, Mao X, et al. Comparison of genetic profiles among primary lung tumor, metastatic lymph nodes and circulating tumor DNA in treatment-naïve advanced non-squamous non-small cell lung cancer patients. Lung Cancer 2018;121:54-60. [Crossref] [PubMed]
- Ou S-HI, Madison R, Robichaux JP, et al. Characterization of 648 non-small cell lung cancer (NSCLC) cases with 28 unique HER2 exon 20 insertions. J Clin Oncol 2019;37:9063. [Crossref]
- Burke CL, Stern DF. Activation of Neu (ErbB-2) mediated by disulfide bond-induced dimerization reveals a receptor tyrosine kinase dimer interface. Mol Cell Biol 1998;18:5371-9. [Crossref] [PubMed]
- He L, Hristova K. Pathogenic Activation of Receptor Tyrosine Kinases in Mammalian Membranes. J Mol Biol 2008;384:1130-42. [Crossref] [PubMed]
- Yamamoto H, Higasa K, Sakaguchi M, et al. Novel Germline Mutation in the Transmembrane Domain of HER2 in Familial Lung Adenocarcinomas. JNCI: Journal of the National Cancer Institute 2014;106:djt338 [Crossref] [PubMed]
- Wang R, Zhang Y, Pan Y, et al. Comprehensive investigation of oncogenic driver mutations in Chinese non-small cell lung cancer patients. Oncotarget 2015;6:34300. [Crossref] [PubMed]
- Oh DY, Bang YJ. HER2-targeted therapies — a role beyond breast cancer. Nat Rev Clin Oncol 2020;17:33-48. [Crossref] [PubMed]
- Baraibar I, Mezquita L, Gil-Bazo I, et al. Novel drugs targeting EGFR and HER2 exon 20 mutations in metastatic NSCLC. Crit Rev Oncol Hematol 2020;148:102906 [Crossref] [PubMed]
- Serra V, Vivancos A, Puente XS, et al. Clinical response to a lapatinib-based therapy for a Li-Fraumeni syndrome patient with a novel HER2V659E mutation. Cancer Discov 2013;3:1238-44. [Crossref] [PubMed]
- Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008;455:1069-75. [Crossref] [PubMed]
- Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012;150:1107-20. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016;48:607-16. [Crossref] [PubMed]
- Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446-51. [Crossref] [PubMed]
- Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2109-21. [Crossref] [PubMed]
- Jordan EJ, Kim HR, Arcila ME, et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer Discov 2017;7:596-609. [Crossref] [PubMed]
- Vavalà T, Monica V, Lo Iacono M, et al. Precision medicine in age-specific non-small-cell-lung-cancer patients: Integrating biomolecular results into clinical practice-A new approach to improve personalized translational research. Lung Cancer 2017;107:84-90. [Crossref] [PubMed]
- Hellmann MD, Nathanson T, Rizvi H, et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell 2018;33:843-52.e4. [Crossref] [PubMed]
- Rizvi H, Sanchez-Vega F, La K, et al. Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J Clin Oncol 2018;36:633-41. [Crossref] [PubMed]
- Chen J, Yang H, Teo ASM, et al. Genomic landscape of lung adenocarcinoma in East Asians. Nat Genet 2020;52:177-86. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data: Figure 1. Cancer Discovery 2012;2:401-4. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Wang Z, Yang JJ, Huang J, et al. Lung Adenocarcinoma Harboring EGFR T790M and In Trans C797S Responds to Combination Therapy of First- and Third-Generation EGFR TKIs and Shifts Allelic Configuration at Resistance. J Thorac Oncol 2017;12:1723-7. [Crossref] [PubMed]
- Sun Y, Ren Y, Fang Z, et al. Lung adenocarcinoma from East Asian never-smokers is a disease largely defined by targetable oncogenic mutant kinases. J Clin Oncol 2010;28:4616-20. [Crossref] [PubMed]
- Pahuja KB, Nguyen TT, Jaiswal BS, et al. Actionable Activating Oncogenic ERBB2/HER2 Transmembrane and Juxtamembrane Domain Mutations. Cancer Cell 2018;34:792-806 e5.
- Morris GM, Huey R, Lindstrom W, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem 2009;30:2785-91. [Crossref] [PubMed]
- Fuhrmann J, Rurainski A, Lenhof HP, et al. A new Lamarckian genetic algorithm for flexible ligand-receptor docking. J Comput Chem 2010;31:1911-8. [Crossref] [PubMed]
- Case DA, Cheatham TE 3rd, Darden T, et al. The Amber biomolecular simulation programs. J Comput Chem 2005;26:1668-88. [Crossref] [PubMed]
- Duan Y, Wu C, Chowdhury S, et al. A Point-Charge Force Field for Molecular Mechanics Simulations of Proteins Based on Condensed-Phase Quantum Mechanical Calculations. J Comput Chem 2003;24:1999-2012. [Crossref] [PubMed]
- Wang J, Wolf RM, Caldwell JW, et al. Development and Testing of a General Amber Force Field. J Comput Chem 2004;25:1157-74. [Crossref] [PubMed]
- Jakalian A, Bush BL, Jack DB, et al. Fast, Efficient Generation of High-Quality Atomic Charges. AM1-BCC Model: I. Method. J Comput Chem 2000;21:132-46. [Crossref]
- Jakalian A, Jack DB, Bayly CI. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J Comput Chem 2002;23:1623-41. [Crossref] [PubMed]
- Wang J, Wang W, Kollman PA, et al. Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graph Model 2006;25:247-60. [Crossref] [PubMed]
- Jorgensen WL, Chandrasekhar J, Madura JD, et al. Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics 1983;79:926-35. [Crossref]
- Kollman PA, Massova I, Reyes C, et al. Calculating Structures and Free Energies of Complex Molecules: Combining Molecular Mechanics and Continuum Models. Acc Chem Res 2000;33:889-97. [Crossref] [PubMed]
- Miller BR, McGee TD, Swails JM, et al. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J Chem Theory Comput 2012;8:3314-21. [Crossref] [PubMed]
- Wilkinson L. ggplot2: elegant graphics for data analysis by WICKHAM H. Biometrics 2011;67:678-9. [Crossref]
- Therneau TM. A Package for Survival Analysis in R. R package version 3.2-7 ed2020.
- Ross JS, Fakih M, Ali SM, et al. Targeting HER2 in colorectal cancer: The landscape of amplification and short variant mutations in ERBB2 and ERBB3. Cancer 2018;124:1358-73. [Crossref] [PubMed]
- Loree JM, Bailey AM, Johnson AM, et al. Molecular Landscape ofERBB2/ERBB3Mutated Colorectal Cancer. J Natl Cancer Inst 2018;110:1409-17. [Crossref] [PubMed]
- Mazieres J, Barlesi F, Filleron T, et al. Lung cancer patients with HER2 mutations treated with chemotherapy and HER2-targeted drugs: results from the European EUHER2 cohort. Ann Oncol 2016;27:281-6. [Crossref] [PubMed]
- Chuang JC, Stehr H, Liang Y, et al. ERBB2-Mutated Metastatic Non-Small Cell Lung Cancer: Response and Resistance to Targeted Therapies. J Thorac Oncol 2017;12:833-42. [Crossref] [PubMed]
- Arcila ME, Chaft JE, Nafa K, et al. Prevalence, clinicopathologic associations, and molecular spectrum of ERBB2 (HER2) tyrosine kinase mutations in lung adenocarcinomas. Clin Cancer Res 2012;18:4910-8. [Crossref] [PubMed]
- Ricciardi GR, Russo A, Franchina T, et al. NSCLC and HER2: between lights and shadows. J Thorac Oncol 2014;9:1750-62. [Crossref] [PubMed]
- Suzawa K, Toyooka S, Sakaguchi M, et al. Antitumor effect of afatinib, as a human epidermal growth factor receptor 2-targeted therapy, in lung cancers harboring HER2 oncogene alterations. Cancer Sci 2016;107:45-52. [Crossref] [PubMed]
- Lai W-CV, Feldman DL, Buonocore DJ, et al. PD-L1 expression, tumor mutation burden and response to immune checkpoint blockade in patients with HER2-mutant lung cancers. J Clin Oncol 2018;36:9060. [Crossref]
- Mazieres J, Drilon A, Lusque A, et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET registry. Ann Oncol 2019;30:1321-8. [Crossref] [PubMed]
- Guisier F, Dubos-Arvis C, Viñas F, et al. Efficacy and Safety of Anti-PD-1 Immunotherapy in Patients With Advanced NSCLC With BRAF, HER2, or MET Mutations or RET Translocation: GFPC 01-2018. J Thorac Oncol 2020;15:628-36. [Crossref] [PubMed]
- Barf T, Kaptein A. Irreversible Protein Kinase Inhibitors: Balancing the Benefits and Risks. J Med Chem 2012;55:6243-62. [Crossref] [PubMed]
- Bauer RA. Covalent inhibitors in drug discovery: from accidental discoveries to avoided liabilities and designed therapies. Drug Discovery Today 2015;20:1061-73. [Crossref] [PubMed]
- Li BT, Li T, Johnson ML, et al. Safety and efficacy of pyrotinib in patients with NSCLC and other advanced solid tumors with activating HER2 alterations: A phase I basket trial. J Clin Oncol 2020;38:3510. [Crossref]
- Wang Y, Jiang T, Qin Z, et al. HER2 exon 20 insertions in non-small-cell lung cancer are sensitive to the irreversible pan-HER receptor tyrosine kinase inhibitor pyrotinib. Ann Oncol 2019;30:447-55. [Crossref] [PubMed]
(English Language Editor: B. Draper)


