
Role of the epithelial-mesenchymal transition-related circular RNA, circ-10720, in non-small-cell lung cancer
Introduction
Worldwide, lung cancer remains the cancer most frequently diagnosed and the leading cause of cancer-related death (1). Lung cancer can be classified in two main histological groups: small-cell lung cancer and non-small-cell lung cancer (NSCLC), which comprises around 85% of all lung cancers (2). The most frequent histological subtypes of NSCLC are adenocarcinoma (ADK), squamous cell carcinoma (SCC) and large cell carcinoma (3,4). Despite the development of novel targeted therapies, NSCLC patients still have a poor prognosis, mainly related to their disease stage at diagnosis. Most patients are diagnosed with advanced or even metastatic disease, while just 16% of all cases are diagnosed at an early stage. This scenario is aggravated by the asymptomatic development of lung cancer, which often presents with non-specific symptoms, such as thoracic pain, cough, dyspnea, or weight loss. Consequently, 5-year survival rates are low (10.7%) (5). In early-stage (I–II) and locally advanced (III) NSCLC, complete surgical resection, when possible, is recognized as a potentially curative treatment. Depending on disease stage, patients can receive adjuvant chemotherapy after surgical resection (6), usually a cisplatin-based combination, which has been proven to confer a benefit in overall survival (7,8). However, 30–70% of patients who undergo surgery will relapse in the first 5 years (9).
Circular RNA (or circRNA) is a type of single-stranded non-coding RNA which forms a covalently closed continuous loop where the 3’ and 5’ ends normally present in RNA molecules have been joined together (10,11). CircRNAs are usually expressed at low levels, but some of them can be more abundant than their linear transcripts. CircRNAs have a high degree of stability since they are resistant to RNAses acting over the linear RNA ends. Their expression is highly tissue-specific and modulates during development or in different cell fates (12). They perform several functions, one of which is the sponging of microRNAs (13-15), where a circRNA sequesters several microRNAs that inhibit the translation of a target gene. This then allows the upregulation of the protein levels of the gene. Other functions of circRNAs have also been described, including sequestering of RNA binding proteins and acting as protein scaffolds (16), and some circRNAs can be translated (17). Different conditions can alter circRNA expression and may affect the expression of its linear form. The deregulation of their levels may have important roles in disease and their detection in human saliva, blood, and exosomes could be explored in the search for new biomarkers (12).
Among the many hallmarks of cancer (18), activating invasion and metastasis is the most critical for patient survival and one in which epithelial-mesenchymal transition (EMT) plays an essential role. EMT is commonly defined by the loss of the epithelial marker E-cadherin (CDH1) and the gain of the mesenchymal marker Vimentin (VIM) (19,20), which is one of the main events in the mesenchymal-like conversion of epithelial cells and one of the best indicators of EMT (21,22). VIM is expressed in epithelial cells when they are involved in physiological or pathological processes requiring epithelial cell migration (23). EMT has been associated with various tumor functions, including tumor initiation, malignant progression, tumor stemness, tumor cell migration, extravasation to the blood, and resistance to therapy (24).
Several studies have shown that circRNAs are frequently produced by back-splicing from a precursor mRNA (pre-mRNA) in specific moments (25). For example, when Quaking protein is upregulated during EMT, it is able to regulate transcription to induce a back-splicing of the pre-mRNA, thus increasing the circRNA concentration (26). In addition, a large number of circRNAs have been related to EMT (15,27). Specifically, in NSCLC, circ_100876 is upregulated in tumor tissue and has been associated with metastasis (28), while circ_0014130 is also upregulated in tumor tissue and is associated with TNM stage and metastasis (29).
Recently, Meng et al. have described a novel circRNA, circ-10720 (CircBase ID: hsa_circ_0018189), derived from the CUL2 gene in hepatocellular carcinoma (HCC). CUL2 acts as a tumor-suppressor protein by regulating angiogenesis and cell cycle. The authors showed that Twist1, which is a mesenchymal factor upregulated in EMT, bound to the CUL2 gene promoter enhancing a back-splicing that ends with the formation of circ-10720 instead of CUL2 mRNA. While CUL2 acted as tumor suppressor, the circ-10720 played an antagonistic role as oncogene and its expression was associated with poor prognosis in HCC (22). Circ-10720 acted as a microRNA sponge for some microRNAs targeting VIM. The upregulation of circ-10720 led to an increase of VIM protein levels by sequestering miR-490-5p, miR-578 and miR-1246, thus activating a mesenchymal phenotype. To date, however, the role of circ-10720 has not been explored in other tumors (30).
We have examined the role of circ-10720 in NSCLC cell lines and patient tumor samples in order to assess its potential use as a biomarker of time to relapse (TTR) in surgically resected NSCLC patients.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-920).
Methods
Cell lines and culture
Cryopreserved samples of the lung cancer cell lines HCC44 (ACC 534, DSMZ, Branuschweig, Germany), A549 (ACC 107, DSMZ), H23 (CRL-5800, ATTC, Manassas, VA, USA) and H1299 (CRL-5803, ATCC); and the normal immortalized lung cell line BEAS2B (95102433, ECACC, Sigma-Aldrich) were received in our laboratory and passaged for less than 6 months. HCC44, H23 and H1299 cell lines were cultured in RPMI 1640 (catalog number 21875091, Thermo Fisher Scientific, Waltham, MA, USA) and A549 was cultured in DMEM medium (catalog number 11995073, Thermo Fisher Scientific, Waltham, MA, USA). All mediums were supplemented with 10% FBS (catalog number 16000044, 1:10, Thermo Fisher Scientific). BEAS2B was cultured using BEGMTM-2 BulletKit Medium (catalog number CC-3170, Lonza, Basel, Basel, Switzerland). All cell lines were grown at 37 °C in 5% CO2 and 95% relative humidity.
RNA extraction and gene expression analysis
Total RNA was isolated from tissue and cell samples using TriZol® Reagent (Thermo Fisher Scientific) following manufacturer’s instructions. RNA was quantified using NanoDrop ND-1000 device (Thermo Fisher Scientific). The reverse transcription reaction was carried out using 500 ng of RNA and the High Capacity cDNA Reverse Transcription Kit® (Thermo Fisher Scientific). The gene expression was determined by Real Time PCR in Step One Real Time PCR System (Thermo Fisher Scientific). Circ-10720 was quantified using a custom PrimeTime Std qPCR Assay (IDT, Coralville, Iowa, USA): primer 1, 5'-GATTTCTCGGAGCAGCATTC-3'; primer 2, 5'-CTGTTTGCAAAGCACCTGAA-3'; Probe/56-FAM/TGATGGTTG/ZEN/AACCACTTCAGGCCA/3IABkFQ/. TaqMan expression assays (Thermo Fisher Scientific) for CUL2 (Hs00180203), VIM (Hs00958111), CDH1 (Hs01023894) and MDM2 (Hs01066930_m1) were used to quantify their expression levels. CDKN1-Β (Hs00153277) was used as endogenous control. Relative expression was calculated using 2–ΔΔCt method.
Cell transfection
On day 0, 1×105 cells were seeded in 12-well plates. On day 1, the cells were transfected with 100 nM of dsiRNA-circ-10720 (custom DsiRNA, IDT) or with Negative Control DsiRNA (IDT) using Lipofectamine 2000 (Thermo Fisher Scientific). An additional validation of the key obtained results were performed using a second dsiRNA, siRNA circ-10720 #2 (Figures S1,S2). The sequences of both dsiRNAs used in the study are provided in Table S1. Circ-10720 inhibition was evaluated for all dsiRNAs by Real Time PCR at 48 hours after transfection.
Western blot
Western blot was performed as previously described by our group (31) using the following primary antibodies: VIM (rabbit polyclonal, dilution 1:3,000, catalog number 10366-1-AP, ProteinTech, Manchester, UK), CDH1 (mouse monoclonal, Clone 36, dilution 1:500, catalog number 610182, BD Transduction Laboratories, San Jose, CA, USA) and α-actin (mouse monoclonal, Clone C4, dilution 1:1,000, catalog number 691001, MP Biomedicals, Solon, OH, USA). Antibody binding was revealed by incubation with anti-mouse (rabbit polyclonal, dilution 1:5,000, catalog number A9044, Sigma-Aldrich, Merk Merck Life Science, Madrid, Spain) or anti-rabbit (goat polyclonal, dilution 1:5,000, catalog number sc-2004, Santa Cruz Biotechnology, Dallas, TX, USA) IgG peroxidase conjugate secondary antibodies. Protein expression was assessed using HPR Chemiluminescent Substrate Reagent Kit (catalog number WP20005, Invitrogen, Thermo Fisher Scientific) and ChemiDoc (BIO-RAD, Hercules, CA, USA).
Immunofluorescence
Cells growing on glass coverslips were processed for immunofluorescence analysis 48 h after transfection with siRNA or control. First, cells were fixed with 2% formaldehyde in PBS for 5 minutes at room temperature (RT). Following fixation, cells were washed in PBS for 10 minutes at RT. Permeabilization of the cells was achieved by incubation for 30 minutes in PBS 0.1% Triton at RT. Then blocking solution [PBS 0.1% Triton, BSA 1% and 5% normal donkey serum (NDS)] was added on top of the coverslip and incubated in a humidified chamber for 1 h. Once blocked, cells were incubated overnight at 4 °C with mouse anti-VIM ready-to-use antibody (mouse monoclonal, clone V9, catalog number IS630, DAKO, Mississauga, Canada). The next day, samples were washed 3 times with PBS during 10 minutes and incubated for 2 hours with goat anti-mouse IgG H&L (Alexa Fluor® 488, catalog number ab150113, Abcam, Cambridge, MA, USA) diluted 1:2,000 in PBS 0.1% Triton, 5% NDS. Finally, samples were washed 3 times with PBS during 5 minutes and mounted on a slide using Fluoroshield mounting medium (Sigma-Aldrich, Merck Life Science) which incorporates DAPI staining and observed under an Olympus BX51 fluorescence microscope.
Wound-healing assay
Cell migration was measured by wound-healing assay. 1.5×105 cells were plated in a 12-well plate and transfected with siRNA or Control at 8 hours. Sixteen hours after transfection, the cell monolayer was scraped in a straight line to create a “scratch” with a p1000 pipet tip. The migration distance (µm) was assessed at different time points until the scratch was closed (24 hours) using cellSense Entry 1.7 software (Olympus, Tokyo, Japan).
Cell invasion assay
Invasion capacity was measured using QCM ECMatrix Cell Invasion Assay (ECM550, Sigma-Aldrich, Merk Merck Life Science) following manufacturer’s instructions. After 24 hours of transfection, 5×105 cells were seeded on the top of the invasion chamber and invasion was quantified at 24 h.
Caspase 3/7 assay
Apoptosis was measured using Caspase-Glo® 3/7 Assay System (Promega, Madison, WI, USA) as per manufacturer’s protocol. Sixteen hours after transfection, 104 cells were seeded on coated 96-well plates. An equal volume of Caspase-Glo 3/7 reagent was added to each well and incubated during 1.30 h at RT. Relative light intensity was measured in each well using a Veritas microplate luminometer (Turner Biosystems, Sunnyvale, CA, USA).
Patient samples
From June 2007 to November 2015, tumor and paired normal tissue were prospectively collected from 119 adult patients diagnosed with NSCLC who underwent complete surgical resection at Hospital Clinic of Barcelona (Spain) and that consented to participate in the study. Samples were immediately immersed in RNALater® (Thermo Fisher Scientific) after collection and stored at –80 °C until processing. None of the patients received neoadjuvant treatment before surgery. Thirty-six (30.3%) received adjuvant cisplatin-based chemotherapy (5 stage I, 18 stage II, 13 stage III) and 83 (69.7%) received no treatment until relapse. Median follow-up time was 61.60 months (IQR: 47.57–88.50). Main characteristics of the 119 patients included in the study are shown in Table 1.
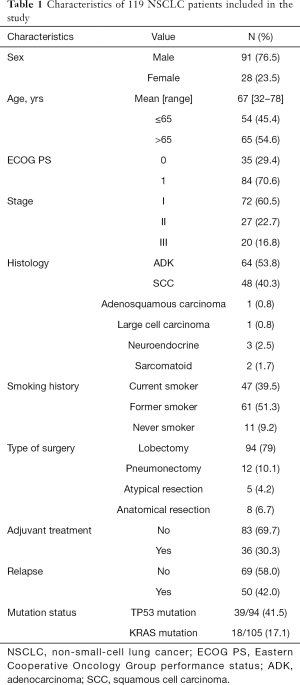
Full table
Statistical analyses
Students t-test was used to compare differences between two groups. At least three technical and three biological replicates have been performed in each in vitro experiment. The primary endpoint analyzed was TTR, calculated from the time of surgery to the date of relapse or last follow-up. Kaplan-Meier curves for TTR were drawn and compared by means of a log-rank test. All factors with a P value <0.1 in the univariate analysis were included in the Cox multivariate regression analyses for TTR. Optimal cutoffs of circ-10720 expression for TTR was identified using X-Tile v.3.6.1 (Yale University 2003–2005) (32). All statistical analyses were performed using IBM SPSS Statistics v25 or GraphPad Prism 8 and R v3.5.1. The level of significance was set at P≤0.05.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Approval for the study was obtained from the center’s institutional review board (ethical approval code HCB/2017/1052), and written informed consent was obtained from each participant.
Results
Circ-10720 regulates VIM in NSCLC cell lines and affects cell morphology
We explored circ-10720 expression in the BEAS2B, a normal immortalized human bronchial epithelial cell line, and in four different NSCLC cell lines, A549, HCC44, H23 and H1299. HCC44 and A549 cell lines overexpressed circ-10720 in comparison with BEAS2B (P=0.035 and P=0.027, respectively), while in H23 and H1299 circ-10720 was downregulated (P=0.011 and P=0.037, respectively; Figure 1A). We also analyzed CUL2 mRNA expression levels and found that its expression in the NSCLC cell lines was not significantly different from that in BEAS2B.

To analyze whether differences in EMT exists between the 4 NSCLC cell lines analyzed we used the EMT markers described in Pastushenko et al. (19), which review the main genes implied in the different transition states from full epithelial to full mesenchymal states. We downloaded the expression data (from RNAseq) of 29 EMT genes from Cancer Cell Line Encyclopedia (CCLE, Broad Institute, https://portals.broadinstitute.org/ccle) (33) and analyzed them using Hierarchical cluster analysis. The Hierarchical cluster analysis showed that the four cell lines grouped based on the EMT profile according to their circ-10720 levels (Figure 1B). This result indicates that differences in the EMT expression profile exists between the A549 and HCC44 (high circ-10720) and H23 and H1299 (low circ-10720) cell lines.
Based on the higher circ-10720 levels HCC44 and A549 cell lines were selected for further study. First, we inhibited circ-10720 expression in both cell lines and found that after 48 h there was a reduction of 80% in HCC44 (P=0.0014) and 70% in A549 (P=0.0067) (Figure 1C). Interestingly, CUL2 mRNA levels were not significantly modified after silencing circ-10720.
We then evaluated the protein levels of VIM and CDH1 in the two cell lines. The silencing of circ-10720 produced a downregulation of VIM in both HCC44 (P=0.036) and A549 (P=0.044). Although not significant, an upregulation of CDH1 could also be observed, especially in the HCC44 (P=0.0726) cell line (Figure 1D,E). However, there were no significant differences in the mRNA expression of either VIM or CDH1 (Figure S3).
Previously (22), it has been reported that VIM regulation of circ-10720 in HCC was mediated by microRNA sponging of miR-490-5p, miR-578 and miR-1246. To test if these microRNAs were involved in VIM regulation in NSCLC cell lines, we analyzed their expression by RT-qPCR. The expression analysis of the three miRNAs in HCC44 and A549 cell lines showed that only miR-1246 was expressed. We could not detect expression of neither miR-490-5p nor miR-578. The analysis of miR-1246 expression after silencing circ-10720 showed an increment in its levels in the silenced group in comparison with the control group of 2.975 folds in HCC44 (P=0.0666) and 3.246 folds in A549 (P=0.0039) of mean, which supports the ceRNA mechanism (Figure 1F).
Interestingly, the changes in VIM protein levels were associated with a modification in the cell morphology. By immunofluorescence using VIM antibody, we observed that silenced cells presented a change in the VIM filament organization in the cells (Figure 1G). These changes in VIM filament organization were associated to cell morphology changes as observed in the bright field images (Figure 1H and Figure S4). Circ-10720 silenced cells showed a more rounder morphology instead of the longer morphology associated with a more mesenchymal state observed in the control cells.
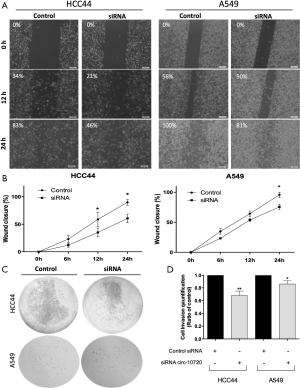
Circ-10720 regulates migration and invasion
We then analyzed the role of circ-10720 on migration and invasion capacity by wound-healing assay and cell invasion assay. Downregulation of circ-10720 was related to a decreased migration capacity (Figure 2A), as shown by a significant difference of wound closure percentage at 24 h (HCC44, P=0.037; A549, P=0.0169; Figure 2B). Moreover, silenced cells had a lower invasion capacity than control cells (HCC44, P=0.0035; A549, P=0.0470) (Figure 2C,D).
Circ-10720 regulates apoptosis and proliferation
To further investigate the role of circ-10720 in NSCLC, we examined the repercussions of its silencing on apoptosis and proliferation. Transfection with siRNA increased apoptosis and reduced proliferation. Analysis at 16 hours after transfection showed that Caspase 3/7 activity was 41% (P=0.0198) and 60% (P=0.0434) higher in HCC44 and A549 respectively, than in control cells (Figure 3A). Proliferation was analyzed at different time points and significant differences were observed for both cell lines. At 72 hours, proliferation was reduced a mean of 37% (P=0.0148) and 64% (P=0.0171) in HCC44 (Figure 3B) and A549 (Figure 3C) respectively.

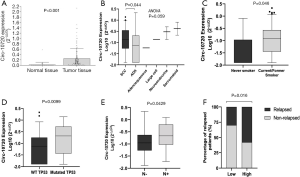
Circ-10720 expression in patient tumor samples
This study was conducted in a cohort of 119 resected NSCLC patients. The main clinical characteristics are summarized in Table 1. Circ-10720 was upregulated in tumor tissue in comparison with normal tissue (P<0.001; Figure 4A). The majority (74%) of normal tissue samples did not show detectable levels of circ-10720. Circ-10720 was differentially expressed in the histological subtypes (ANOVA P=0.059; Figure 4B) and was overexpressed in SCC compared to ADK (P=0.044). In addition, circ-10720 was expressed at higher levels in current and former smokers than in never smokers (P=0.046; Figure 4C). Patients harboring TP53 mutations also had higher levels of circ-10720 than those with wild-type TP53 (P=0.0099; Figure 4D). Interestingly, circ-10720 expression, was significantly higher in patients with lymph node involvement (N+, n=16) (P=0.0429, Figure 4E). Moreover, patients with higher levels had higher relapse rate (Figure 4F).
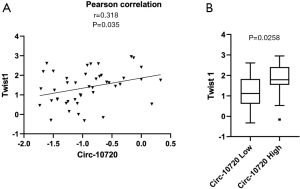
Circ-10720 expression in patient tumor samples correlates with Twist1 expression
We have analyzed the potential correlation of circ-10720 expression with the expression of different EMT markers in the tumor tissue of 40 NSCLC patients of our cohort. The following EMT markers have been analyzed: Twist1, CDH1, VIM, ZEB1, ZEB2 and SNAIL. The analysis of these six EMT-related genes showed that circ-10720 levels positively correlated with Twist1 (r=0.318; P=0.035, Figure 5A). Of note, in the 40 samples analyzed Twist1 were positively correlated with ZEB1 (r=0.383; P=0.0101). Interestingly, when the patients were classified in high vs. low circ-10720 expression according to the prognostic cutoff, we observed that patients with high circRNA levels had higher levels of Twist1 (P=0.0258) (Figure 5B).
Circ-10720 is associated with TTR in patients not receiving adjuvant treatment
In the entire cohort of 119 patients, those with high levels of circ-10720 had shorter median TTR than those with low levels (89.7 months vs. not reached; P=0.293; Figure S5). Although this difference was not statistically significant for the entire cohort, when we evaluated the relapse rate in the subgroup of 83 patients not receiving adjuvant treatment (main clinical characteristics summarized in Table S2), we found that 58% of those with high circ-10720 levels relapsed, compared to 30% of those with low levels (P=0.016; Figure 4F). In non-treated patients, median TTR for patients with high circ-10720 expression was 61 months (95% CI: 7–115) and was not reached for those with high levels (P=0.027; Figure 6).
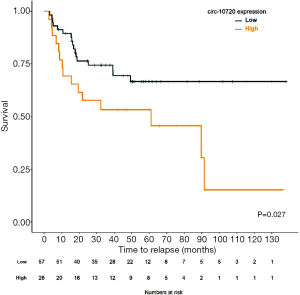
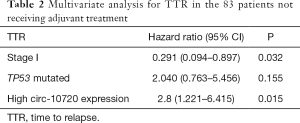
Then, circ-10720 expression, disease stage, and TP53 mutational status were included in the multivariate analyses of TTR in patients not receiving adjuvant treatment (Table 2). High circ-10720 expression (HR: 2.8; 95% CI: 1.221–6.415; P=0.015) emerged as an independent prognostic marker for shorter TTR, while stage I disease (HR: 0.291; 95% CI: 0.094–0.897; P=0.032) was an independent marker for longer TTR.
Full table
Discussion
In the present study, we analyzed the role of circ-10720 in NSCLC and its potential as a biomarker of relapse after surgery in resected stage I–IIIA NSCLC patients. We found that modulation of circ-10720 levels regulates EMT, apoptosis and proliferation in vitro and correlated with EMT markers and prognosis in patient samples in NSCLC.
We first quantified the expression of circ-10720 in four NSCLC cell lines and in a normal immortalized lung cell line, BEAS2B, and observed an inter-cell line variability. HCC44 and A549 cell lines had higher levels, while H23 and H1299 had low levels in comparison to the BEAS2B cells. We did not observe significant differences in the expression of the CUL2 gene.
We explored whether the differences in circ-10720 levels observed in the four NSCLC cell lines where related to different EMT stage. For this we used the EMT markers described in Pastushenko et al. (19), which review the main genes implied in the different transition states, allowing the classification of samples in five main stages: epithelial tumor cells, early hybrid EMT state, hybrid EMT state, late hybrid EMT state and mesenchymal tumor cell. The hierarchical cluster analysis showed that the four cell lines grouped based on the EMT profile according to their circ-10720 levels indicating that differences in the EMT expression profile exists between the A549 and HCC44 (high circ-10720) and H23 and H1299 (low circ-10720) cell lines. We performed a preliminary analysis to try to identify the EMT genes differentially expressed between the two groups. This analysis had the limitation of having only two cell lines (two values) per group (a t-test with only one degree of freedom) and have to be considered a primary approximation and for this we have not included the results of this analysis in the main text, but in our opinion deserves to be discussed. The identification of differentially expressed genes showed that KRT14, PRRX1, and ASPN were differentially expressed between both groups (high vs. low circ-10720 expression). KRT14 is an epithelial marker that is not expressed in the mesenchymal tumor cell state (can be found in epithelial, early hybrid EMT and late hybrid EMT states), while PRRX1 and ASPN are only expressed in mesenchymal tumor cell state. The two cell lines with low levels of circ-10720 had lower levels of KRT14, higher levels of PRRX1 and expressed ASPN indicating that they are in a complete mesenchymal state. We can speculate that circ-10720 levels are higher in cells that have not completed the EMT transition, which is indicated by the expression of genes such as PRRX1 and ASPN. Moreover, we observed that HCC44 and A549 were in different EMT stages as shown by the different genes included in the analysis, which seems to indicate that A549 is in a more advanced EMT stage (higher VIM and lower CDH1 and EPCAM levels).
We then focused on functional studies in the two cell lines overexpressing circ-10720, where we inhibited circRNA levels using a siRNA. Since in HCC the oncogenic role of this circRNA has been related to EMT (22), we performed several in vitro assays to evaluate if it also regulated EMT in NSCLC. An efficient inhibition of circ-10720 in both cell lines was associated with lower VIM protein levels and a trend towards higher CDH1 levels. Moreover, morphological changes were observed in both cell lines after circRNA silencing, with loss of the mesenchymal phenotype, indicating that circ-10720 silencing could be associated with mesenchymal to epithelial transition (MET). During EMT, epithelial characteristics are lost, while mesenchymal features are temporarily acquired. This process is marked by increasing levels of VIM, which allows the activation of cellular processes such as cell mobility, invasiveness, or reinforced resistance to apoptosis (20,34). After migration, cells may reverse their phenotype through MET (35). Reduced aggressiveness of tumor cells in terms of migration and invasion capacity were also observed after circ-10720 silencing. The wound-healing assay and the invasion assay showed that migration was reduced by 32% and 21%, while invasion was reduced by 18% and 21% in HCC44 and A549, respectively. These results are in line with those by Meng et al., who showed that circ-10720 regulated EMT in HCC through regulation of VIM levels by sponging several microRNAs targeting VIM, including miR-490-5p, miR-578 and miR-1246 (22). However, we observed that only miR-1246 was expressed in A549 and HCC44 cell lines. Moreover, the silencing of circ-10720 correlated with an increase of the levels of miR-1246 suggesting that is involved in the ceRNA mechanism of VIM regulation. MiR-1246 have been previously involved in NSCLC initiation (36) and metastasis trough regulation of EMT (37).
Additionally, since reinforced resistance to apoptosis is a typical feature acquired during EMT, we examined the impact of circ-10720 inhibition on apoptosis (38). The inhibited cells presented a higher activation of Caspase 3/7 than control cells, which is a direct measure of apoptosis, indicating a role for circ-10720 in apoptosis regulation. Moreover, we could confirm that this had an impact on proliferation by MTS analysis from 0 to 72 h. This effect on apoptosis and proliferation regulation could be due to different mechanisms, including direct and indirect regulation mediated by the circRNA. circ-10720 has been shown to have other microRNA targets, some of which could regulate apoptosis-related genes. We analyzed using Diana-miRPath (39) the pathways affected by the microRNA set validated to bind circ-10720 (22) and identified 13 significant KEGG pathways associated with them, including the p53 signaling pathway, where 14 genes were regulated by four microRNAs binding the circRNA. One of the relevant genes identified was MDM2, which was regulated by miR-331-5p. Moreover, an additional analysis using miRWalk (40) identified other microRNAs from the set also regulating MDM2, including miR-890, miR-127-5p and miR-578. Using in silico data from TCGA with TACCO database (41), we identified a negative correlation between MDM2 mRNA levels and miR-127-5p. When we explored the mRNA levels of MDM2 after circ-10720 silencing, we observed that at least in one cell line (A549), the MDM2 mRNA levels were significantly reduced up to 19% in the silenced group (data not shown). However, further investigation is warranted to explore the role of circ-10720 and its regulated microRNAs in apoptosis regulation, since this is beyond the scope of the present paper.
Based on the results obtained in relation to apoptosis and proliferation, we could discuss whether the migration results obtained are a consequence of the increased cell death and slowed growth. However, we think that the invasion assay confirmed that circ-10720 regulates cell mobility apart from proliferation and apoptosis. Of note, the regulation of invasion was more marked in the HCC44 cell line than in the A549 cell line. Then we could speculate than the migration effect observed in A549 cell line is probably more related to the regulation of proliferation and apoptosis in this cell line than in HCC44. Nevertheless, both effects, mobility and growth can be modulated at the same time during EMT.
Based on our finding that circ-10720 had a role in EMT and apoptosis regulation in NSCLC cell lines, we analyzed its expression in 119 patient tumor and normal tissue samples. Although circ-10720 expression was low in general, there was a clear upregulation in tumor tissue, while in 74% of normal tissues, its expression was not detectable. Moreover, it was differentially expressed in different histological subtypes, with the highest levels in sarcomatoid, neuroendocrine and large cell carcinoma. This is in line with a previous report that these groups express higher levels of VIM (23). However, SCC had higher levels than ADK, which cannot be explained by VIM levels since ADK has higher levels of VIM than SCC (42). We observed upregulation of circ-10720 in current and former smokers and in TP53-mutated patients. Some tobacco compounds have been associated with the induction of EMT in lung cancer (43), making tobacco a potential inductor of circ-10720 expression. Moreover, along these same lines, upregulation of VIM has also been previously observed in smokers (44). The upregulation of circ-10720 in TP53-mutated patients may be explained by a recent bioinformatic analysis showing a potential association between TP53 mutations and increased circRNA levels (45).
Moreover, we explored whether circ-10720 levels correlated with the EMT status of tumor tissues. For this, we analyzed the expression of several EMT markers in a subset of 40 tumor tissues of the cohort. The analysis showed that circ-10720 levels positively correlated with Twist1, a typical mesenchymal marker. Twist1 has been shown to be involved in the back splicing related to circ-10720 production and its expression was also correlated with circ-10720 levels in HCC (22). Of note, we observed that Twist1 were positively correlated with ZEB1 reinforcing the idea that high circ-10720 correlates with a more mesenchymal phenotype in patient tumor samples.
Finally, we explored the role of circ-10720 expression as a potential biomarker of relapse in NSCLC and found that its prognostic impact was restricted to the 82 patients who had not received adjuvant chemotherapy after surgery, mainly stage I patients (81%). Moreover, 9 stage II (10.8%) and 7 stage III (8.5%) patients also not received adjuvant treatment and were included in the analysis. The main causes that explained why this group of stage II–III patients not received adjuvant treatment were: (I) own patient decision; or (II) Unfit patient. In this subset of patients, those with higher expression levels of circ-10720 had a higher relapse rate and shorter TTR than those with lower levels. Moreover, circ-10720 emerged as an independent prognostic marker for TTR in the multivariate analysis. Although the prognostic role of circ-10720 has not been previously described in NSCLC, the prognostic impact of VIM levels has been widely studied. In our in vitro study, we verified that in NSCLC—as in HCC—circ-10720 silencing reduces VIM levels and enhances MET. Overexpression of VIM has been associated with poor prognosis in NSCLC patients (23). A prognostic impact for circ-10720 levels was also described in HCC (22). Other EMT-related circRNAs have been associated with poor prognosis in NSCLC. For example, shorter overall survival was associated with high circ_0020123 (46), and circ_100876 (28) levels. Our findings in the subset of patients not receiving treatment after surgery suggest that adjuvant treatment might well prevent relapse in patients with high circ-10720 levels.
To the best of our knowledge, this is the first study to report a prognostic impact for circ-10720 in NSCLC. However, we need to discuss some limitations of the present study. Although the exact mechanism of circ-10720 action in NSCLC is not fully addressed in the present study, our results indicate that circ-10720 expression regulate EMT, apoptosis and proliferation in vitro. In vivo studies to confirm the role of circ-10720 in the regulation of metastasis process will deserve further investigation and will further verify the conclusions of the present study. Moreover, we showed that circ-10720 may be a biomarker of risk of relapse and their expression correlate with a more mesenchymal phenotype. However, further investigation in a prospective study or in independent cohorts is warranted to validate these findings and to examine potential circ-10720-based therapeutic approaches.
Conclusions
Circ-10720 in NSCLC play a role as an oncogene in NSCLC regulating migration, invasion, apoptosis and proliferation in vitro. Circ-10720 regulation of migration and invasion is performed trough regulation of EMT by modification of VIM levels. Moreover, their levels in tumor samples correlate with mesenchymal markers and acts as relapse biomarker in non-treated early-stage NSCLC patients.
Acknowledgments
Funding: This work was supported by grants from the Ministry of Economy, Industry, and Competition, Agencia Estatal de Investigación co-financed with the European Union FEDER funds SAF2017-88606-P (AEI/FEDER, UE) and SDCSD from the Universitat de Barcelona.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-920
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-920
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-920). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Institutional Review Board of Hospital Clínic of Barcelona (ethical approval code HCB/2017/1052), and written informed consent was obtained from each participant in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Oberndorfer F, Mullauer L. Molecular pathology of lung cancer: current status and perspectives. Curr Opin Oncol 2018;30:69-76. [Crossref] [PubMed]
- Testa U, Castelli G, Pelosi E. Lung cancers: molecular characterization, clonal heterogeneity and evolution, and cancer stem cells. Cancers (Basel) 2018;10:248. [Crossref] [PubMed]
- Chansky K, Detterbeck FC, Nicholson AG, et al. The IASLC Lung Cancer Staging Project: external validation of the revision of the TNM stage groupings in the eighth edition of the TNM classification of lung cancer. J Thorac Oncol 2017;12:1109-21.
- Chirlaque MD, Salmeron D, Galceran J, et al. Cancer survival in adult patients in Spain. Results from nine population-based cancer registries. Clin Transl Oncol 2018;20:201-11. [Crossref] [PubMed]
- Wu AJ, Garay E, Foster A, et al. Definitive radiotherapy for local recurrence of NSCLC after surgery. Clin Lung Cancer 2017;18:e161-8. [Crossref] [PubMed]
- Bessho Y, Oguri T, Ozasa H, et al. ABCC10/MRP7 is associated with vinorelbine resistance in non-small cell lung cancer. Oncol Rep 2009;21:263-8. [PubMed]
- Postmus PE, Kerr KM, Oudkerk M, et al. Early and locally advanced non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2017;28:iv1-21. [Crossref] [PubMed]
- Felip E, Rosell R, Maestre JA, et al. Preoperative chemotherapy plus surgery versus surgery plus adjuvant chemotherapy versus surgery alone in early-stage non-small-cell lung cancer. J Clin Oncol 2010;28:3138-45. [Crossref] [PubMed]
- Ebbesen KK, Hansen TB, Kjems J. Insights into circular RNA biology. RNA Biol 2017;14:1035-45. [Crossref] [PubMed]
- Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science 2013;340:440-1. [Crossref] [PubMed]
- Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol 2018;11:98. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol 2016;238:42-51. [Crossref] [PubMed]
- Du WW, Yang W, Liu E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 2016;44:2846-58. [Crossref] [PubMed]
- Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell 2017;66:9-21.e7. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol 2019;29:212-26. [Crossref] [PubMed]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest 2009;119:1420-8. [Crossref] [PubMed]
- Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest 2009;119:1429-37. [Crossref] [PubMed]
- Meng J, Chen S, Han JX, et al. Twist1 Regulates vimentin through Cul2 circular RNA to promote EMT in hepatocellular carcinoma. Cancer Res 2018;78:4150-62. [Crossref] [PubMed]
- Dauphin M, Barbe C, Lemaire S, et al. Vimentin expression predicts the occurrence of metastases in non small cell lung carcinomas. Lung Cancer 2013;81:117-22. [Crossref] [PubMed]
- Flier SN, Tanjore H, Kokkotou EG, et al. Identification of epithelial to mesenchymal transition as a novel source of fibroblasts in intestinal fibrosis. J Biol Chem 2010;285:20202-12. [Crossref] [PubMed]
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453-61. [Crossref] [PubMed]
- Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015;160:1125-34. [Crossref] [PubMed]
- Lv J, Liu J, Guo L, et al. Bioinformatic analyses of microRNA-targeted genes and microarray-identified genes correlated with Barrett's esophagus. Cell Cycle 2018;17:792-800. [Crossref] [PubMed]
- Yao JT, Zhao SH, Liu QP, et al. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract 2017;213:453-6. [Crossref] [PubMed]
- Zhang S, Zeng X, Ding T, et al. Microarray profile of circular RNAs identifies hsa_circ_0014130 as a new circular RNA biomarker in non-small cell lung cancer. Sci Rep 2018;8:2878. [Crossref] [PubMed]
- Navarro A. Twist1 activated circRNA-10720 is a new player in hepatocellular carcinoma metastasis. Transl Cancer Res 2018;78:4150-62.
- Cordeiro A, Navarro A, Gaya A, et al. PiwiRNA-651 as marker of treatment response and survival in classical Hodgkin lymphoma. Oncotarget 2016;7:46002. [Crossref] [PubMed]
- Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 2004;10:7252-9. [Crossref] [PubMed]
- Ghandi M, Huang FW, Jané-Valbuena J, et al. Next-generation characterization of the cancer cell line encyclopedia. Nature 2019;569:503-8. [Crossref] [PubMed]
- Tejero R, Navarro A, Campayo M, et al. miR-141 and miR-200c as markers of overall survival in early stage non-small cell lung cancer adenocarcinoma. PLoS One 2014;9:e101899 [Crossref] [PubMed]
- Thompson EW, Haviv I. The social aspects of EMT-MET plasticity. Nat Med 2011;17:1048-9. [Crossref] [PubMed]
- Zhang WC, Chin TM, Yang H, et al. Tumour-initiating cell-specific miR-1246 and miR-1290 expression converge to promote non-small cell lung cancer progression. Nat Commun 2016;7:11702. [Crossref] [PubMed]
- Xu X, Cao L, Zhang Y, et al. MicroRNA-1246 inhibits cell invasion and epithelial mesenchymal transition process by targeting CXCR4 in lung cancer cells. Cancer Biomark 2018;21:251-60. [Crossref] [PubMed]
- Yochum ZA, Burns TF. TWIST1 regulation of circRNA: a novel mechanism to promote epithelial-mesenchymal transition in hepatocellular carcinoma. Noncoding RNA Investig 2018;2:71. [Crossref] [PubMed]
- Vlachos IS, Zagganas K, Paraskevopoulou MD, et al. DIANA-miRPath v3. 0: deciphering microRNA function with experimental support. Nucleic Acids Res 2015;43:W460-6 [Crossref] [PubMed]
- Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods 2015;12:697. [Crossref] [PubMed]
- Chou PH, Liao WC, Tsai KW, et al. tACCo, a database connecting transcriptome alterations, pathway alterations and clinical outcomes in Cancers. Sci Rep 2019;9:3877. [Crossref] [PubMed]
- Upton MP, Hirohashi S, Tome Y, et al. Expression of vimentin in surgically resected adenocarcinomas and large cell carcinomas of lung. Am J Surg Pathol 1986;10:560-7. [Crossref] [PubMed]
- Vu T, Jin L, Datta PK. Effect of cigarette smoking on epithelial to mesenchymal transition (EMT) in lung cancer. J Clin Med 2016;5:44. [Crossref] [PubMed]
- Ye Z, Zhang X, Luo Y, et al. Prognostic values of vimentin expression and its clinicopathological significance in non-small cell lung cancer: a meta-analysis of observational studies with 4118 cases. PLoS One 2016;11:e0163162 [Crossref] [PubMed]
- Yu Z, Hou Y, Wang R, et al. TP53 mutations lead to increased transcription of circular rnas in exosomes enhancing tumor invasiveness in hepatocellular carcinoma bases on bioinformatics analysis. Am J Transplant 2018;18:719-20.
- Qu D, Yan B, Xin R, et al. A novel circular RNA hsa_circ_0020123 exerts oncogenic properties through suppression of miR-144 in non-small cell lung cancer. Am J Cancer Res 2018;8:1387-402. [PubMed]


