
Presentation of EGFR mutations in 162 family probands with multiple primary lung cancer
Introduction
Multiple primary lung cancer (MPLC) refers to the synchronous or metachronous occurrence of two or more primary lung cancers in the same patient (1). With advances in chest computed tomography (CT) imaging in recent years, the number of MPLC patients has rapidly increased (2-4). MPLC is relatively common among female nonsmokers and has an excellent prognosis (3). The incidence of first-degree relatives of MPLC patients with a family history of this disease is significantly higher (5). Our previous study also revealed that the incidence of malignant tumors in first-degree relatives of MPLC patients was as high as 35.8% (102/285) (6). A recent study also suggested that MPLC has unique genetic characteristics and should therefore be treated as an independent disease (4). All the above findings suggested a potentially different etiology for MPLC; however, etiological studies of MPLC are still rare.
Epidermal growth factor receptor (EGFR) germline mutations, including the mutations p.T790M (7-9) and p.R776H (10,11) in exon 20 and p.V843I in exon 21 (12,13), are associated with genetic susceptibility to lung cancer. The EGFR-T790M germline mutation was first identified in a European family with a history of MPLC and was associated with drug resistance (14). Several other studies reported the existence of the EGFR-T790M germline mutation in familial MPLC cases (14,15). Our previous study also revealed that the EGFR-R776H germline mutation existed in a family with a high incidence of MPLC (11). These studies have facilitated the understanding of how germline mutations in EGFR may regulate the occurrence of MPLC. The few described cases to date have been insufficient to uncover the genetic relationship between EGFR mutation and MPLC due to the low frequency of MPLC patients with mutant EGFR.
Lung cancers with specific somatic EGFR mutations have a favorable clinical response to EGFR-tyrosine kinase inhibitors (TKIs) (16-18). The most frequent EGFR mutations are small insertions and deletions (indels) around exon 19 and the p.L858R mutation in exon 21, which are oncogenic and hypersensitive to EGFR-TKIs. Other less prevalent but clinically relevant EGFR mutations that predict a resistant response to EGFR-TKIs include p.G719A/S/C in exon 18; p.dels in exon 19; p.S768I, p.T790M, and p.indels in exon 20; and p.L861Q in exon 21 (19).
To identify the signatures of candidate germline and tumor-derived mutations in family members and to provide evidence of etiology and clinical TKI treatment for high-risk MPLC patients, 162 probands [162 noncancerous adjacent tissues (NATs) and 366 tumor lesions] were screened in the current study. Additionally, somatic mutations were described in the other 3,243 sporadic lung adenocarcinoma (LUAD) patient cohorts. We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-1001).
Methods
Study design and patient selection
In all, 366 tumor lesions and 162 NATs from a cohort of 162 family probands were separately genotyped for candidate germline mutations and somatic mutations (Figure 1A). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). Ethical approval was obtained from the medical ethics committee of the National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (NCC2018AA-015). And all the patients provided written informed consent. As a control cohort for somatic mutations in the current study, samples from 3243 LUAD patients were previously subjected to ten-gene target sequencing (20). All EGFR mutant sites were described in detail in both cohorts (Figure 1B).
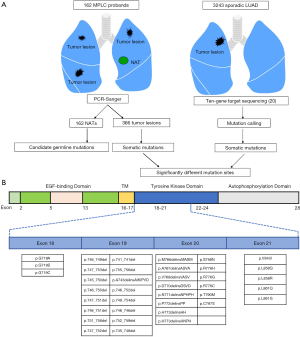
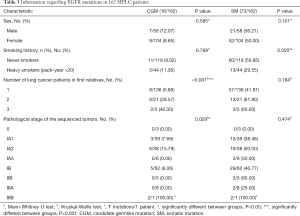
Selection criteria included a diagnosis of MPLC and sufficient archived tissue samples available for PCR sequencing. Each proband represents one family with at least one affected first-degree relative with MPLC, and a total of 162 probands were analyzed in our study. The study population was limited to surgically treated family probands with pathological diagnosis of MPLC at our hospital between January 2011 and January 2017. The inclusion criteria for patients were as follows: (I) pathologically confirmed MPLC (according to the 8th American Joint Committee on Cancer (AJCC) edition; typical CT and pathological images of MPLC are shown in Figure 2) and at least one first-degree relative with pathologically confirmed primary lung cancer; and (II) available complete medical records. The source for detecting EGFR mutations was formalin-fixed, paraffin-embedded (FFPE) normal tissues and tumor tissues that were collected after resection. Pathological diagnosis and staging (according to the 8th AJCC edition) were performed by two pathologists and two thoracic surgeons, respectively, who were blinded to the patient data. Detailed characteristics of the 3243 patients with sporadic LUAD were described in a previous study by the current authors (20).
FFPE tissue preparation and DNA extraction
Lung tumor and adjacent normal tissue samples were formalin fixed and paraffin embedded after surgical resection to produce FFPE samples. Genomic DNA (gDNA) was extracted from the FFPE samples using the GeneRead DNA FFPE Kit (Qiagen, Hilden, Germany) and quantified with Qubit (Life Technologies, Carlsbad, California, USA) in accordance with the manufacturer’s protocol.
EGFR mutation detection in 162 MPLC patients
All mutations were amplified from the gDNA of 162 probands using standard PCR. PCR was conducted using the following gene-specific sense and antisense primers: exon 18: F (5′-GAGGTGACCCTTGTCTCTGTGT-3′) and R (5′-CCCAAACACTCAGTGAAACAAA-3′), exon 19: F (5′-CAATATCAGCCTTAGGTGCGG-3′) and R (5′-GATGTGGAGATGAGCAGGGTC-3′), exon 20: F (5′-GTCCATGTGCCCCTCCTTCT-3′) and R (5′-TTCCCTGATTACCTTTGCGA-3′), and exon 21: F (5′-CGTTCGCCAGCCATAAGTCCT-3′) and R (5′-GCATCCTCCCCTGCATGTGTT-3′). PCR amplification and agarose gel detection were repeated 3 times, and two researchers independently interpreted the mutation data and gave reports.
EGFR somatic mutation analysis in 3,243 patients with sporadic LUAD
Sequencing data for 3,243 patients with sporadic LUAD from Geneis (Beijing) Co., Ltd. were read using Finch TV software (Geospiza Inc., Seattle, USA), and genotyping was performed. Variant calling was performed on the Lung Cancer Ten-Genes Panel (Geneis Co., Ltd.) from NextSeq CN500 sequencing with Burrows-Wheeler Aligner (BWA) and FreeBayes software. The common clinical databases used in this study included PharmGKB, the Human Gene Mutation Database (HGMD), Clinvar, Catalogue of Somatic Mutations in Cancer (COSMIC), SNPedia, 1000 Genomes and dbSNP (20). A blinded approach was followed using frequency thresholds of ≥1% and ≥5% to define a mutation for the tumor tissue samples analyzed.
Statistical analysis
SPSS version 13.0 (IBM Corporation, Chicago, IL, USA) was used. Intergroup differences were compared using the chi-square test or Fisher’s exact test. Mann-Whitney U test and Kruskal-Wallis test were used for analysis of continuous variables with a non-normal distribution respectively. A value of P<0.05 was considered significant.
Results
Patients and samples
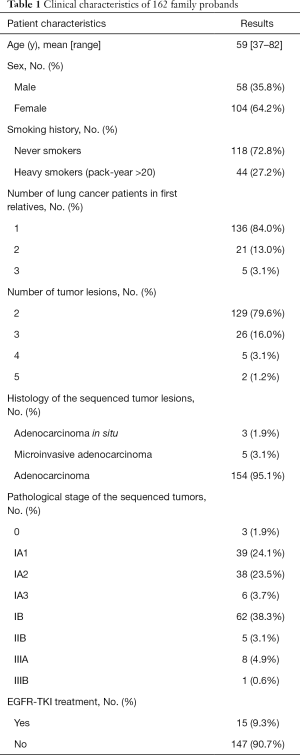
A total of 162 MPLC probands with 366 tumor lesions and 162 corresponding nontumor normal tissue samples were screened for EGFR mutations. Samples were obtained from paraffin-embedded tissues deposited in a tumor bank. The clinicopathological characteristics of the 162 probands are summarized in Table 1. We chose multiple lesion samples and corresponding nontumor normal tissue samples for PCR sequencing.
Full table
Detailed characteristics of the 3,243 patients with sporadic LUAD were described in a previous study by the current authors (20).
Distribution of candidate germline EGFR mutations in 162 MPLC patients
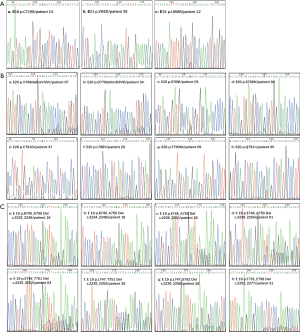
We proposed that EGFR mutations are common variants that predispose individuals to lung cancer and that they are potentially enriched in probands representing lung cancer families. gDNA from 162 MPLC family probands was analyzed. PCR amplification and DNA sequencing of EGFR exon 18 p.G719X, 19 p.dels, 20 p.S768I, 20 p.R776H/G/C, 20 p.T790M, 20 p.C797S, 20 p.indels (p.M766indelsMASM, p.A767indelsASVA, p.V769indelsVASV, p.D770indelsDSVD, p.N771indelsNPHPH, p.P772indelsPP, p.H773indelsHH, and p.H773indelsHNPH), 21 p.V843I, 21 p.L858R and 21 p.L861Q/G were performed (Figure 3A,B,C). The mutations described above were detected in 162 nontumor tissue samples adjacent to cancerous tissue with the following distribution: exon 19, p.746_750del (2, 1.23%) and p.746_751del (1, 0.62); exon 20, p.V769indelsVASV (2, 1.23%), p.S768I (3, 1.85%), p.S768N (1,0.62%), p.T790M (1, 0.62%), and p.Q791* (1, 0.62%); and exon 21, p.V843I (1, 0.62%) and p.L858R (4, 2.74%) (Figure 3B,C, Table 2 and Table S1). As shown in Table 2, the total frequency of mutations in nontumor tissues was 9.88%.
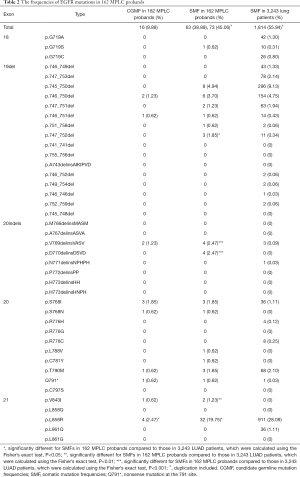
Full table
Distribution of somatic EGFR mutations in 162 MPLC patients
PCR sequencing of 366 tumor lesion samples for somatic EGFR driver mutations was also carried out. The results showed 74 different heterozygous mutations, including exon 18 p.G719S (1, 0.62%), 19 p.dels (21, 12.35%), 20 p.insX (8, 4.94%), 20 p.S768I (3, 1.85%), 20 p.S768N (1, 0.62%), 20 p.C781Y (1, 0.62%), 20 p.L788V (1, 0.62%), 20 p.T790M (3, 1.85%), 20 p.Q791* (1, 0.62%), 21 p.V843I (2, 1.23%) and 21 p.L858R (32, 19.75%) (Figure 3A,B,C and Table 2). Of the 74 mutations, multiple mutations were observed in eight patients, and repeated counting was avoided. A total of 63 MPLC patients among initial 162 (38.89%) were identified as having somatic mutations (duplications removed) (Table S1). We performed a stratified analysis to assess sex, smoking status, number of lung cancer patients in first relatives and pathological stage. Results showed smoking status impacted on somatic mutant rate (P<0.05, Table 3 and Table S2).
Full table
The detailed information for the patients with coexisting mutations was as follows: patient 20 (exon 19 p.745_750del and 20 p.L788V), patient 24 (exon 19 p.746_750del and 21 p.L858R), patient 1 (exon 19 p.746_750del and 20 p.S768I), patient 3 (exon 19 p.746_751del, 20 p.T790M and 21 p.L858R), patient 7 (exon 20 p.V769indelsVASV and 21 p.L858R), patient 4 (exon 20 p.V769indelsVASV, 20 p.S768N and 21 p.L858R), patient 5 (exon 20 p.S768I and 20 p.Q791*) and patient 9 (exon 20 p.T790M and 21 p.L858R) (Table S1). We also analyzed the mutation presentation in the 366 tumor lesion samples. The results showed low consistency between lesions individually, and only 9 cases showed consistency (Tables S1,S2).
Comparison of EGFR somatic mutations in 162 MPLC probands and in 3243 LUAD patients
In total, 3243 sporadic LUAD tissue samples were previously subjected to targeted sequencing using the lung cancer ten-gene panel (Geneis Co., Ltd.). We reanalyzed the data and identified the EGFR somatic mutation frequencies (threshold of ≥1%) of exon 18 p.G719X (78, 2.41%), 19 p.dels (668, 20.60%), 20 p.indels (4, 0.12%), 20 p.S768I (36, 1.11%), 20 p.R776H/G/C (12, 0.37%), 20 p.T790M (68, 2.10%), 21 p.L858R (911, 28.06%) and 21 p.L861Q (36,1.11%). These results are summarized in Table 2. First, as the most common mutations, the frequencies of exon 19 p.dels and 21 p.L858R mutation were indeed the highest in the two cohorts. However, there were significant differences in several mutations: exon 19 p.747-752del, 20 p.V769indelsVASV, 20 p.D770indelsDSVD, 21 p.V843I and 21 p.L858R. Specifically, the chi-square test showed the exon 19 p.747_752del, 20 p.V769indelsVASV, 20 p.D770indelsDSVD and 21 p.V843I mutations occurred at significantly higher frequencies in the MPLC population (1.85%, 2.47%, 2.47% and 1.23%, respectively) than in the LUAD population (0.34%, 0.09%, and 0.00%, respectively; P value <0.05) (Figure S1).
Discussion
Lung cancer is a major cause of death in China and other countries. Genetic factors play a major role in the development of lung cancer, especially familial lung cancer (21). Although there have been many studies addressing the molecular characteristics of MPLC in recent years, the number of cases involved in these studies has not been higher than 100 (4,22,23). Moreover, only several case reports have documented germline mutations in MPLC (9-13). Our study, for the first time, reported candidate germline mutations and tumor somatic mutations in a large-scale cohort of MPLC probands. In all, 162 patients with MPLCs and a first-degree relative with lung cancer were identified, which is a very impressive cohort size that can only be achieved at a high-volume thoracic surgical center in a region with a high incidence of EGFR mutant lung cancer.
Germline mutations in EGFR are rare but may contribute to oncogenesis (7-13,24-26). A larger study on the p.T790M germline mutation in 31,414 patients reported that 48 patients (0.15%) carried the germline p.T790M mutation (27). This mutation is one of the best studied germline mutations, which has been previously estimated to be involved in 1% to 4% of EGFR-positive lung cancers and implicated in hereditary lung cancer (28). The exon 20 p.R776H germline mutation is of special concern in MPLC patients according to our previous results (11); interestingly, the p.R776H/G/C mutation was not detected in any of the family probands analyzed in this study. Here, we systematically investigated the signature of EGFR mutations in a population of MPLC families. The most important result was that several candidate germline mutations with frequencies above 1% were detected. These candidate germline mutations are as follows: exon 19 p.746_750del (2, 1.23%), 20 p.V769indelsVASV (2, 1.23%), 20 p.S768I (3, 1.85%), and 21 p.L858R (4, 2.74%). Exon 20 exhibited the highest frequency of mutations in NATs (8/16, 50%). Exon 20 encodes the third most predominant kinase domain, and the mutation class encompasses in-frame insertions and indels following or within the regulatory C-helix amino acids (29,30). These oncogene variants engage the phosphoinositide 3-kinase/protein kinase B and extracellular-signal-regulated kinase/mitogen-activated protein kinase downstream pathways with EGFR dependency (30). Although the EGFR p.S768I mutation is considered to be highly rare, we detected it in a total of 1.85% (3/162) of the probands with MPLC. It is worth emphasizing that all p.S768I mutations identified were candidate germline mutations. All of the above mentioned candidate germline mutations might be responsible for the etiology of MPLC.
Predictive biomarker testing for EGFR point mutations and indels is the standard of care in NSCLC patients for the selection of an optimal first-line therapy (31,32). However, for MPLC patients, an EGFR mutation spectrum to guide clinical medication is lacking. In our study, 162 probands with 366 tumor lesions were screened for EGFR somatic mutations. This study is the first to report exon 18 p.G719S (1, 0.62%); exon 19 p.dels (21, 12.35%); exon 20 p.ins (8, 4.94%), p.S768I (3, 1.85%), p.S768N (1, 0.62%), p.C781Y (1, 0.62%), p.L788V (1, 0.62%), p.T790M (3, 1.85%) and p.Q791* (1, 0.62%); and exon 21 p.V843I (2, 1.23%) and p.L858R (32, 19.75%) mutations in family numbers with MPLC. Exon 19 deletions and exon 21 p.L858R were most common in the MPLC family population. Genetic heterogeneity is a significant molecular genetic feature of MPLC. High-throughput sequencing analysis has previously shown high heterogeneity among multiple lesions in the same MPLC patient (4,33,34). Heterogeneity was also detected in our population, and only 9 patients in the 162 MPLC probands showed consistency between tumor lesions.
In China and other Asian countries, female never-smokers with an EGFR mutation account for a large percentage (40–50%) of LUAD cases (35-38). More than 60% of females and nonsmokers were in our cohort, and the total EGFR mutation rate was 45.06% (73/163) (duplicates included) (Table 2). Smoking status and gender are important factors for EGFR mutation patterns, and we performed a stratified analysis to assess sex, smoking status, number of lung cancer patients in first relatives and pathological stage. We found no difference in smoking status between the two cohorts, and the proportion of female patients in the MPLC cohort was significantly higher (Table S3). However, whether gender differences are the fundamental cause of higher EGFR mutations remains to be further verified. In addition, it is interesting that only some specific site mutation frequencies (Figure S1) rather than all sites in the MPLC population are higher (Table 2), suggesting that they may have more value in clinical applications.
In the current study, we found that the frequencies of exon 19 p.747-752del, 20 p.V769indelsVASV, 20 p.D770indelsDSVD and 21 p.V843I were significantly higher in the MPLC probands (Figure S1). We performed functional prediction with SIFT and PROVEAN, and the results showed that the mutated sites were probably damaging/deleterious (Table S4). Nine nucleotide insertion mutations (p.V769indelsVASV and p.D770indelsDSVD) in exon 20 of EGFR were detected in eight patients with MPLC (Figure 3B). The frequencies of exon 20 p.indels were 4.94% in our family population, 0.09% in the 3243 patients with sporadic LUAD and 0.31% (88/28,768) in the COSMIC database. This implies that these mutations might be related to the occurrence of familial MPLC. The effect of EGFR-TKIs has been examined in cultured cell lines transfected with exon 20 insertion mutations. The 50% inhibitory concentration (IC50) of osimertinib was lower than that of other EGFR-TKIs in all cell lines tested (39). Clinical trials are currently underway to examine the effects of several EGFR-TKIs on NSCLC with confirmed exon 20 insertion mutations (UMIN000031929, ClinicalTrials.gov Identifier: NCT03066206, and ClinicalTrials.gov Identifier: NCT03414814). We hope that these results will establish evidence that osimertinib and other EGFR-TKIs are effective for NSCLC positive for exon 20 insertion mutations.
There are several limitations to our study. First, the distribution of EGFR mutations in the 162 MPLC cases was identified by PCR-based Sanger sequencing; however, the 3243 sporadic adenocarcinoma samples had been characterized by next-generation sequencing (NGS) with a ten-gene panel. The PCR-Sanger is highly effective and accurate for sequencing single targeted genes and a single reaction can easily yield DNA sequence data between 500 and 1,000 base pairs in length. One major issue of this technology can not suitable for detecting low-frequency mutations for low sensitivity (40). EGFR has a high-frequency mutation in Chinese, and our sample size is relatively large (162 probands). NGS has advantages in high sensitivity and multiplexed testing. However, the quality of the results is dependent on sample type, method, sequencing platform and bioinformatic pipeline and finally interpretation by a team of molecular and genetic experts (40). The most important results in the study is that the rate of EGFR mutations in exon 19 p.747-752del, 20 p.V769indelsVASV and 20 p.D770indelsDSVD were significantly higher in MPLC probands detected with sanger than those in sporadic LUAD patients detected with NGS. For the low sensitivity of sanger sequencing compared with NGS, this result gives more stronger evidence that mutations in exon 19 p.747-752del, 20 p.V769indelsVASV and 20 p.D770indelsDSVD may paly significant roles in family MPLC. Second, candidate germline mutations were identified in normal tissue adjacent to cancerous tissue. A recent study emphasized that blood is superior to these adjacent normal tissues (41), but NATs were used for identification of candidate germline mutations in our study due to the lack of peripheral blood samples. Third, the frequency of EGFR mutation in NSCLC has been described to differ across ethnic groups and the occurrence was shown to be evidently higher in East-Asians compared with that in Europeans (42,43). the two cohorts involved in the study are all from East Asian populations, and this conclusion should be carefully interpreted when applied to other ethnicities.
Conclusions
This study first showed the unique EGFR mutation signatures in MPLC probands. We also first screened high-frequency EGFR candidate germline mutations, such as exon 19 p.del, 20 p.ins, 20 p.S768X and 21 p.L861R. Compared with the LUAD patients, MPLC probands had a high frequency of pathogenic somatic mutations, including exon 19 p.747-752del and 20 p.V769indelsVASV and p.D770indelsDSVD. These findings might provide evidence that relates to the etiology of these mutations and shed light on the clinical treatment of high-risk patients.
Acknowledgments
Funding: This work was supported by the National Key R&D Program of China (2018YFC1312100), the National Natural Science Foundation of China (81972196), The CAMS Innovation Fund for Medical Sciences (CIFMS) (2017-I2M-1-005, 2019-I2M-2-002), The Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2018PT32033), The Innovation team development project of Ministry of Education (IRT_17R10).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-1001
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-1001
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-1001). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). Ethical approval was obtained from the medical ethics committee of the National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (NCC2018AA-015). And all the patients provided written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Martini N, Melamed MR. Multiple primary lung cancers. J Thorac Cardiovasc Surg 1975;70:606-12. [Crossref] [PubMed]
- Jung EJ, Lee JH, Jeon K, et al. Treatment outcomes for patients with synchronous multiple primary non-small cell lung cancer. Lung Cancer 2011;73:237-42. [Crossref] [PubMed]
- Detterbeck FC, Bolejack V, Arenberg DA, et al. The IASLC Lung Cancer Staging Project: Background Data and Proposals for the Classification of Lung Cancer with Separate Tumor Nodules in the Forthcoming Eighth Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2016;11:681-92.
- Park E, Ahn S, Kim H, et al. Targeted Sequencing Analysis of Pulmonary Adenocarcinoma with Multiple Synchronous Ground-Glass/Lepidic Nodules. J Thorac Oncol 2018;13:1776-83. [Crossref] [PubMed]
- Li X, Hemminki K. Familial multiple primary lung cancers: a population-based analysis from Sweden. Lung Cancer 2005;47:301-7. [Crossref] [PubMed]
- Zhang Z, Gao S, Mao Y, et al. Surgical Outcomes of Synchronous Multiple Primary Non-Small Cell Lung Cancers. Sci Rep 2016;6:23252. [Crossref] [PubMed]
- Prudkin L, Tang X, Wistuba II. Germ-line and somatic presentations of the EGFR T790M mutation in lung cancer. J Thorac Oncol 2009;4:139-41. [Crossref] [PubMed]
- Tibaldi C, Giovannetti E, Vasile E, et al. Inherited germline T790M mutation and somatic epidermal growth factor receptor mutations in non-small cell lung cancer patients. J Thorac Oncol 2011;6:395-6. [Crossref] [PubMed]
- Oxnard GR, Miller VA, Robson ME, et al. Screening for germline EGFR T790M mutations through lung cancer genotyping. J Thorac Oncol 2012;7:1049-52. [Crossref] [PubMed]
- van Noesel J, van der Ven WH, van Os TA, et al. Activating germline R776H mutation in the epidermal growth factor receptor associated with lung cancer with squamous differentiation. J Clin Oncol 2013;31:e161-4. [Crossref] [PubMed]
- Su K, Gao S, Ying J, et al. Sequencing a super multiple synchronous lung cancer reveals a novel variant in driver gene ARID1B. J Thorac Cardiovasc Surg 2018;155:e185-91. [Crossref] [PubMed]
- Ikeda K, Nomori H, Mori T, et al. Novel germline mutation: EGFR V843I in patient with multiple lung adenocarcinomas and family members with lung cancer. Ann Thorac Surg 2008;85:1430-2. [Crossref] [PubMed]
- Ohtsuka K, Ohnishi H, Kurai D, et al. Familial lung adenocarcinoma caused by the EGFR V843I germ-line mutation. J Clin Oncol 2011;29:e191-2. [Crossref] [PubMed]
- Bell DW, Gore I, Okimoto RA, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet 2005;37:1315-6. [Crossref] [PubMed]
- Lou Y, Pecot CV, Tran HT, et al. Germline Mutation of T790M and Dual/Multiple EGFR Mutations in Patients With Lung Adenocarcinoma. Clin Lung Cancer 2016;17:e5-11. [Crossref] [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. [Crossref] [PubMed]
- Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A 2004;101:13306-11. [Crossref] [PubMed]
- Costa DB. Kinase inhibitor-responsive genotypes in EGFR mutated lung adenocarcinomas: moving past common point mutations or indels into uncommon kinase domain duplications and rearrangements. Transl Lung Cancer Res 2016;5:331-7. [Crossref] [PubMed]
- Meng H, Guo X, Sun D, et al. Genomic Profiling of Driver Gene Mutations in Chinese Patients With Non-Small Cell Lung Cancer. Front Genet 2019;10:1008. [Crossref] [PubMed]
- Tomoshige K, Matsumoto K, Tsuchiya T, et al. Germline mutations causing familial lung cancer. J Hum Genet 2015;60:597-603. [Crossref] [PubMed]
- Chung JH, Choe G, Jheon S, et al. Epidermal growth factor receptor mutation and pathologic-radiologic correlation between multiple lung nodules with ground-glass opacity differentiates multicentric origin from intrapulmonary spread. J Thorac Oncol 2009;4:1490-5. [Crossref] [PubMed]
- Asmar R, Sonett JR, Singh G, et al. Use of Oncogenic Driver Mutations in Staging of Multiple Primary Lung Carcinomas: A Single-Center Experience. J Thorac Oncol 2017;12:1524-35. [Crossref] [PubMed]
- Thomas A, Xi L, Carter CA, et al. Concurrent molecular alterations in tumors with germ line epidermal growth factor receptor T790M mutations. Clin Lung Cancer 2013;14:452-6. [Crossref] [PubMed]
- Gazdar A, Robinson L, Oliver D, et al. Hereditary lung cancer syndrome targets never smokers with germline EGFR gene T790M mutations. J Thorac Oncol 2014;9:456-63. [Crossref] [PubMed]
- Girard N, Lou E, Azzoli CG, et al. Analysis of genetic variants in never-smokers with lung cancer facilitated by an Internet-based blood collection protocol: a preliminary report. Clin Cancer Res 2010;16:755-63. [Crossref] [PubMed]
- Hu Y, Alden RS, Odegaard JI, et al. Discrimination of Germline EGFR T790M Mutations in Plasma Cell-Free DNA Allows Study of Prevalence Across 31,414 Cancer Patients. Clin Cancer Res 2017;23:7351-9. [Crossref] [PubMed]
- Gaughan EM, Cryer SK, Yeap BY, et al. Family history of lung cancer in never smokers with non-small-cell lung cancer and its association with tumors harboring EGFR mutations. Lung Cancer 2013;79:193-7. [Crossref] [PubMed]
- Yasuda H, Kobayashi S, Costa DB. EGFR exon 20 insertion mutations in non-small-cell lung cancer: preclinical data and clinical implications. Lancet Oncol 2012;13:e23-31. [Crossref] [PubMed]
- Yasuda H, Park E, Yun CH, et al. Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci Transl Med 2013;5:216ra177 [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Thorac Oncol 2013;8:823-59. [Crossref] [PubMed]
- Leighl NB, Rekhtman N, Biermann WA, et al. Molecular testing for selection of patients with lung cancer for epidermal growth factor receptor and anaplastic lymphoma kinase tyrosine kinase inhibitors: American Society of Clinical Oncology endorsement of the College of American Pathologists/International Association for the study of lung cancer/association for molecular pathology guideline. J Clin Oncol 2014;32:3673-9. [Crossref] [PubMed]
- Wu C, Zhao C, Yang Y, et al. High Discrepancy of Driver Mutations in Patients with NSCLC and Synchronous Multiple Lung Ground-Glass Nodules. J Thorac Oncol 2015;10:778-83. [Crossref] [PubMed]
- Liu Y, Zhang J, Li L, et al. Genomic heterogeneity of multiple synchronous lung cancer. Nat Commun 2016;7:13200. [Crossref] [PubMed]
- Chen J, Yang H, Teo ASM, et al. Genomic landscape of lung adenocarcinoma in East Asians. Nat Genet 2020;52:177-86. [Crossref] [PubMed]
- Nahar R, Zhai W, Zhang T, et al. Elucidating the genomic architecture of Asian EGFR-mutant lung adenocarcinoma through multi-region exome sequencing. Nat Commun 2018;9:216. [Crossref] [PubMed]
- Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers--a different disease. Nat Rev Cancer 2007;7:778-90. [Crossref] [PubMed]
- Wang C, Yin R, Dai J, et al. Whole-genome sequencing reveals genomic signatures associated with the inflammatory microenvironments in Chinese NSCLC patients. Nat Commun 2018;9:2054. [Crossref] [PubMed]
- Hirano T, Yasuda H, Tani T, et al. In vitro modeling to determine mutation specificity of EGFR tyrosine kinase inhibitors against clinically relevant EGFR mutants in non-small-cell lung cancer. Oncotarget 2015;6:38789-803. [Crossref] [PubMed]
- Sridhar K, Singh A, Butzmann A, et al. Molecular genetic testing methodologies in hematopoietic diseases: current and future methods. Int J Lab Hematol 2019;41:102-16. [Crossref] [PubMed]
- Yizhak K, Aguet F, Kim J, et al. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science 2019;364:eaaw0726 [Crossref] [PubMed]
- Smits AJ, Kummer JA, Hinrichs JW, et al. EGFR and KRAS mutations in lung carcinomas in the Dutch population: increased EGFR mutation frequency in malignant pleural effusion of lung adenocarcinoma. Cell Oncol (Dordr) 2012;35:189-96. [Crossref] [PubMed]
- Soh J, Toyooka S, Matsuo K, et al. Ethnicity affects EGFR and KRAS gene alterations of lung adenocarcinoma. Oncol Lett 2015;10:1775-82. [Crossref] [PubMed]


