
Targeting histone deacetylase enhances the therapeutic effect of Erastin-induced ferroptosis in EGFR-activating mutant lung adenocarcinoma
Introduction
Lung cancer is a malignant tumor with the highest mortality, and lung adenocarcinoma (LUAD) accounts for 40% of all lung cancer cases (1). In Asia, epidermal growth factor receptor (EGFR) is the most common driver mutation in LUAD, with an incidence of 55% (2-4). Of those, EGFR-activating mutation in LUAD accounts for 17.4% worldwide and 37.3% in China (5). EGFR tyrosine kinase inhibitors (EGFR-TKIs) currently represent the standard first-line treatment for patients with EGFR mutant LUAD (6). Although using EGFR-TKI has resulted in significant clinical benefit and an unprecedented increase in survival in patients with advanced EGFR mutant NSCLC (7-10), acquired resistance invariably develops. Secondary EGFR mutations, including EGFR-T790M mutations and other mutations within the EGFR domain, mutations in MAPK, PI3K and cell cycle genes and amplification of EGFR or other oncogenes, like MET, lead to acquired EGFR-TKI resistance of LUAD cells (11-13). However, some patients acquired resistance to EGFR-TKI in the absence of known resistance mechanisms. Thus, intrinsic EGFR-TKI resistance is a clinical challenge. It is reported that about 20–30% EGFR-mutant LUAD were intrinsic resistant to EGFR-TKI (14). Therefore, how to overcome these acquired and intrinsic EGFR-TKI resistance has been the focus of clinical attention.
Ferroptosis is a newly discovered form of cell death caused by an accumulation of toxic lipid peroxides (15). Recent evidence suggested that ferroptosis is commonly dysregulated and contributes to tumorigenesis (16), highlighting anti-ferroptosis as a promising anticancer strategy (17) or to overcome therapeutic resistance (18). Lipoxygenases (LOXs) are a set of non-heme iron-containing enzymes that catalyze the peroxidation of free and esterified polyunsaturated fatty acids to corresponding lipid peroxides (19,20) while GPX4 reduces the toxic lipid peroxides to the nontoxic fatty alcohols (19,21). The use of GPX4 inhibitors, including rat sarcoma viral oncogene homolog (RAS)-selective lethal factor 3 (RSL3), can effectively induce ferroptosis (19). GPX4 reduces lipid peroxides using GSH as a cofactor (20). Cysteine is necessary for the synthesis of GSH, and cystine, the precursor of intracellular cysteine, is imported by the amino acid transporter system xc−. system xc– is a heterodimer consisting a light chain (xCT, SLC7A11) and a heavy chain (4F2hc, SLC3A2). xCT is the key protein of the ferroptosis regulatory network (20,22). Overexpression of xCT can protect tumor cells from ferroptosis (23). Dixon et al. (19,20) showed that using xCT inhibitor Erastin may directly inhibit the system xc– activity, reduce cystine uptake and induce ferroptosis. However, Erastin can induce the compensatory increase of xCT (16). Bersuker et al. (24) displayed that ferroptosis inhibitor protein 1 (FSP1), also known as apoptosis-inducing factor mitochondrial associated protein 2 (AIFM2), may inhibit cell ferroptosis through GPX4-independent pathways.
Interestingly, a growing body of evidence has shown that EGFR signaling pathway participates in ferroptosis regulation (25-29). However, it is still unclear whether ferroptosis induction can be used as a new strategy to treat EGFR mutant LUAD or overcome the acquired and intrinsic EGFR-TKI resistance. Therefore, in this study, we attempted to treat intrinsic and acquired EGFR-TKI resistant LUAD by inducing ferroptosis.
The development of therapeutic resistance is common. To further dissect the therapeutic effect of ferroptosis inducers, we searched for targeted drugs that could affect tumor cells’ ferroptosis sensitivity. Our previous studies have found that various potential drugs can induce ferroptosis of cancer cells (30).
In this study, by combining weighted gene co-expression network analysis (WGCNA) with Connectivity Map analysis (CMap), we found that a histone deacetylase inhibitor (HDACi), Vorinostat, could reverse the ferroptosis resistance of tumor cells and further promote the therapeutic effect of ferroptosis induction. Mechanistically, we found that HDACi exerted its role via downregulating SLC7A11 (xCT). Together, in this study, we found that after developing resistance to EGFR-TKIs, EGFR-mut lung cancer cells display hypersensitivity to ferroptosis inducers, which, to our best knowledge, has not been reported. Also, we implemented bioinformatic strategies to search for drugs (HDAC inhibitors) that have the most potential to enhance the effect of ferroptosis inducers. Finally, we confirmed that combined HDAC inhibitors could dramatically enhance the efficacy of ferroptosis inducers. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tlcr-21-303).
Methods
Materials
The materials used in this study are described in Table 1.
Full table
Study methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study research did not involve human or animals, thus ethical approval was waived.
Human LUAD cell lines and cell culture
EGFR mutant LUAD cell lines, HCC827, HCC4006, H1975, H1650, PC9, HCC4011 and H1993, were preserved by our laboratory. Osimertinib resistant cell lines HCC827R and H1975R were established and preserved in our laboratory, according to our previous study (31). The development of resistance to EGFR-TKI in HCC827 (HCC827R) and H1975 (H1975R) was confirmed by cell viability assay (Figure S1A,B). Cells were cultured in a complete medium holding 90% RPMI-1640 medium, 10% fetal bovine serum (FBS), and 1% 100× penicillin and streptomycin, at 5% CO2 and 37 °C incubators.
Detecting ferroptosis of LUAD cells by C11-BODIPY
H1650, HCC827R, H1975R cells were cultured in 6-well plates with 105 cells per well. The next day, H1650 cells were treated with DMSO, Vorinostat 1 µM, Erastin 1 µM, or Erastin 1 µM + Vorinostat 1 µM for 48 h. HCC827R cells were treated with DMSO, Vorinostat 0.5 µM, Erastin 5 µM, or Erastin 5 µM + Vorinostat 0.5 µM for 24 h. C11-BODIPY lipid peroxide probe solution (2.5 µM, using RPMI-1640 medium as solute) was added to each well. After incubation at 37 °C for 30 min, each group’s cells were collected. The level of lipid peroxidation was quantified by flow cytometry (30). The concentration of Erastin and Vorinostat was determined based on the previous literature (16,17,19,21,32,33).
Detecting apoptosis in LUAD cells by Annexin V/PI double staining
H1650 cells were cultured in 6-well plates at 1×105 cells per well. After being cultured overnight, H1650 cells were divided into four groups: control, Vorinostat 1 µM, Erastin 1 µM, Erastin 1 µM + Vorinostat 1 µM. After 48 hours, the cells were collected, incubated with PI and AnnexinV-FITC (34-36). The positive cells for apoptosis (AnnexinV-FITC positive, PI negative, or APC annexin V positive, PI positive) were analyzed by flow cytometry.
Western blotting (WB) analysis
HCC827, HCC4006, H1650, PC9, H1993 and HCC827R cells were cultured in 6 cm dishes and divided into control group, Erastin (1 µM) group, Vorinostat group (1 µM), and combination group (1 µM Vorinostat and 1 µM Erastin). After 24 h treatment, the cells were lysed with RIPA buffer, and the lysate proteins were separated on 10% SDS-PAGE gel. The protein was transferred to the PVDF 0.2 µm membrane by wet transfer. Samples were transferred to the PVDF membrane (Millipore Corporation, IPVH00010), sealed with TBST holding 5% skim milk powder, and incubated for 1 hour at room temperature; the primary antibody was incubated at 4 °C overnight. The membranes were then washed 3× with TBS/T, incubated with secondary antibody at 1:5,000 for 1 hour in TBS/T, and washed 3× with TBS/T. HRP-conjugated bands were imaged using enhanced chemiluminescence reagent (ECL, Thermo Fisher), and chemiluminescence was recorded with aby BIO-RAD ChemiDoc-XRS+ chemiluminescence imager.
Cell counting assay and cell viability assay.
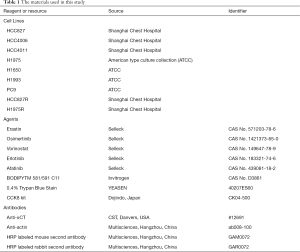
HCC827, HCC4006, H1650, PC9, H1993, H1975, HCC4011, H1975R and HCC827R were inoculated into 6-well plates and divided into control group, Erastin (1 µM) group, Vorinostat group (1 µM) and combination group (1 µM Vorinostat and 1 µM Erastin). After 48 hours of drug treatment, the cells of each group were collected. After trypan blue staining, the living cells in each group were counted. The cell viability was calculated according to the formula: cell viability (%) = [(the number of living cells in the control group) − (the number of living cells in the treated group)]/(the number of living cells in the control group) ×100%. The dose of Erastin and RSL3 that we used to treat EGFR mutant LUAD cells (H1650, H1993, and HCC4011) in Figure 1A,B,C,D,E,F,G,H was lower than the conventional experimental dose (Erastin, 5 µM; RSL3, 1 uM) (16,17,19,20).
Cell proliferation assays
H1650, HCC4011, H1993, H1975R and HCC827R cell lines were seeded at 5×103 cells/well in 96-well plates. Three replicate wells were used for each analysis. After the cells were incubated with different concentrations of drugs for 4 days, respectively, cell viability was monitored with CCK8 kit (CCK8, Dojindo, Japan) following the producer’s suggestions; 10 µL CCK-8 solution was added to each well, incubated for 2 h, and then assayed using a microplate reader with a wavelength of 450 nm. The growth curve was drawn with drug concentrations as the abscissa and absorbance value as the vertical axis.
Data sources and analysis
From the attachment of the literature DOI: 10.1038/nchembio.1986, we obtained the area under the concentration-effect curve (AUC) of Erastin, RSL3, ML162 to 659 kinds of tumor cell lines (37).
We obtained the 659 tumor cell lines, the mRNA expression matrix from the CCLE database (Cancer Cell Line Encyclopedia, https://portals.broadinstitute.org/ccle/about), including 18,543 genes.
WGCNA
We used the R package “WGCNA” for WGCNA analysis to find the key gene modules and hub genes related to ferroptosis sensitivity (32,38,39). According to the dissimilarity measurement for Topological Overlap Matrix (TOM), genes are divided into different gene modules. Here, we set the soft threshold to 4 (scale-free R2=0.96), the cutting height to 0.25, and the minimum number of module genes to 30 to find key modules. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed on the module with the highest correlation with R software’s ferroptosis sensitivity. The genes with GS (gene significance) >0.3 and MM (module membership) >0.8 are defined as hub genes. The co-expression network of 30 hub genes with the highest connectivity was visualized by Cytoscape software (version 3.4.0).
Gene functional enrichment analysis and protein interaction network analysis
The R packet “clusterprofiler” was used for GO enrichment and KEGG pathway enrichment analysis. The GO terms and KEGG pathways with a corrected P value <0.05 were considered significant and were visualized by “GOplot” (R packet). Protein interaction network analysis was performed using STRING (https://string-db.org).
Screening of drugs that promote ferroptosis by using the Connectivity Map database
The top 500 high connectivity genes in the gene modules which was the most positively correlated with ferroptosis sensitivity in WGCNA analysis were taken as a down-tag gene set, and the top 500 high connectivity genes in the gene modules which was the most negatively correlated with ferroptosis sensitivity in WGCNA analysis were taken as an up-tag gene set. The expression profiles of these 1,000 genes were uploaded into CMap (http://www.broad.mit.edu/cmap/) database to obtain the drugs that have the potential to reverse the ferroptosis resistance genotype (gene expression profile) (40).
Statistical analysis
Data were presented as mean ± standard deviation, with the indicated sample size (n) representing biological replicates. Data analysis was performed using R (version 3.6.3) (41-43). Statistical significance was determined by one-way/two-way analysis of variance (ANOVA), Bonferroni’s multiple comparison test, and Student’s t-test using R, unless otherwise indicated. P<0.05 was considered statistically significant.
Results
EGFR-TKI intrinsic resistant LUAD cells and EGFR-TKI acquired resistant LUAD cells are sensitive to ferroptosis inducers
Firstly, we observed that ferroptosis inducers Erastin and RSL3 could significantly decrease cell number of three EGFR mutant LUAD cell lines (H1650, H1993, and HCC4011) (Figure 1A,B). Assessing EGFR-TKI sensitivity, H1650 showed strong resistance to both the first and second generations of EGFR-TKI, followed by H1993, and HCC4011 was the most sensitive (Figure 1C,D). Assessing ferroptosis inducers sensitivity, we could see that H1650, which was the most resistant to EGFR-TKI, was the most sensitive to Erastin and RSL3. However, HCC4011, which was the most sensitive to the first- and second-generation EGFR-TKI, was the most resistant to Erastin and RSL3 (Figure 1E,F). In addition, the lung cancer cell line 1975R, which was resistant to the third-generation EGFR-TKI osimertinib, was more sensitive to ferroptosis inducers than the primary cell line H1975 (Figure 1G,H).
Screening of key genes affecting ferroptosis resistance of tumor cells by WGCNA analysis combined with Connectivity Map technique
The sensitivity of each tumor to ferroptosis was measured by the concentration-effect curve (AUC) of three ferroptosis inducers: Erastin, RSL3, ML162. The mRNA expression matrix (including 18,543 genes of 659 tumor cell lines) and AUC matrix were used to perform WGCNA analysis (Figure 2A). By setting the soft threshold power as 5 (R2=0.92) and the cutting height as 0.25, we identified nine gene modules (Figure 2B,C,D,E, unclustered DEG was shown in gray). From the heatmap of module-trait correlations, we found that the blue module was the most positively correlated with the AUC score of the three ferroptosis inducers, and the black module was the most negatively correlated with the AUC score of the three ferroptosis inducers. By calculating the correlation between the GS and MM, we further verified the genes in the blue module, and the black module was significantly correlated with the ferroptosis sensitivity (Figure S2A,B). The relationship between the genes in the blue module and the blue module and ferroptosis resistance was further verified.
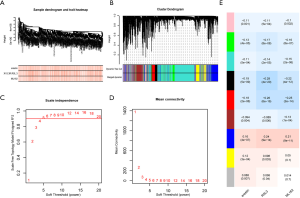
Thus, the overexpression of genes in the blue module could inhibit tumor cells’ ferroptosis, while the overexpression of genes in the black module could promote tumor cells’ ferroptosis. Targeting genes in the blue module could reverse the ferroptosis resistance of tumors.
Co-expression network analysis showed that the expression of 30 most connected genes in the blue and black module was significantly positively correlated (Figure S2C,D) and there was the interaction between the proteins expressed by these genes (Figure S2E,F). We performed GO and KEGG analysis to reveal genes’ potential biological function in the black and blue modules. In GO analysis, the blue module was significantly enriched in epidermal development and epidermal differentiation (Figure 3A,B), while in KEGG enrichment analysis, the blue module was significantly enriched in Wnt and Hippo signal pathways. In GO analysis, the black module was significantly enriched in small GTP enzyme binding and RasGTP enzyme binding, while in KEGG analysis, the black module was significantly enriched in the chemokine-related pathway and Rap1 signal pathway (Figure 3C,D).

Screening drugs that can reverse ferroptosis resistance of EGFR mutant LUAD cells
The top 500 genes with the highest connectivity in the black module were taken as the down-tag gene set, and the top 500 genes with the highest connectivity in the blue module were taken as the up-tag gene set. The 1,000 genes were uploaded into the Connectivity Map (CMap, http://www.broad.mit.edu/cmap/) analysis in QuerySignature file format. The outputs of CMap analysis were sorted according to the mean of the correlation coefficient, and the drugs with negative correlation were proposed to reverse ferroptosis resistance status (40). We took P<0.05 as the criterion for judging the significant effect of drugs, and a total of 33 drugs were selected (Figure 4A). Among the 33 drugs, Vorinostat had the highest specificity (specificity =0.60, Figure 4B). It was suggested Vorinostat could reverse ferroptosis resistance and enhance the therapeutic effect of ferroptosis inducer.
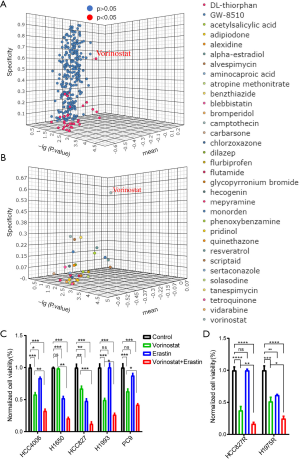
To evaluate this hypothesis, we used Vorinostat and Erastin alone or in combination to treat EGFR mutant LUAD cells (HCC827, HCC4006, PC-9, H1650, H1993). We selected HCC827, HCC4006 and PC9 cells as EGFR TKI sensitive lines and H1650, PC9 cells as EGFR-TKI resistant lines according to the previous studies (44-47). The results showed that the cell numbers of combination group were more prominently diminished than that of single-drug group (Figure 4C). We also observed that Vorinostat significantly enhanced the inhibitory effect of Erastin on the proliferation of EGFR-TKI acquired resistant cell line HCC827R and H1975R (Figure 4D).
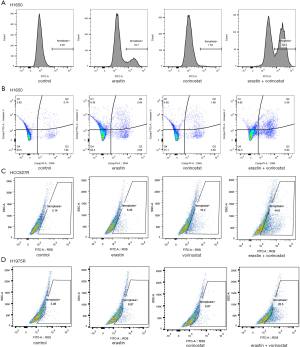
In addition, in the EGFR-TKI resistant H1650 cell line, we found that the level of lipid peroxide, a biomarker of ferroptosis, was higher in the Vorinostat and Erastin combination treatment group than that in the single-drug group (Figure 5A). In addition, apoptosis level of combination treatment group was more obvious than that of single-drug group (Figure 5B). Therefore, we believed that Vorinostat significantly promoted the Erastin-induced ferroptosis of EGFR mutant LUAD cells. Lipid peroxide detection assay also showed that Vorinostat could significantly enhance the Erastin-induced ferroptosis of HCC827R and H1975R cells (Figure 5C,D). It is worth noting that growing evidence in the literature suggests monotherapy with Vorinostat could overcome EGFR-TKI acquired resistance (48).
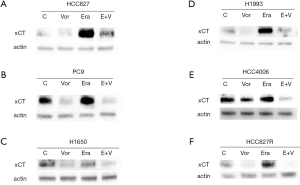
Vorinostat enhances the ferroptosis of EGFR mutant LUAD cells induced by Erastin by down-regulating the expression of xCT
Western blot results showed the expression of xCT protein decreased in HCC827, HCC4006, PC-9, H1650, and H1993 cells after treating them with Vorinostat alone, while the expression of xCT protein increased in these cells after treating them with Erastin alone, while Erastin combined with Vorinostat reversed the compensatory increase of xCT induced by Erastin (Figure 6A,B,C,D,E). Not only that, but we also found comparable results in EGFR-TKI acquired resistant cell line HCC827R (Figure 6F).
Discussion
EGFR is the most frequent oncogenic driver mutation in Chinese patients with LUAD (2-4). The standard first-line treatment for LUAD with EGFR mutation includes EGFR-TKI (49). Although most patients with LUAD harboring EGFR-TKI-sensitizing mutations have an initial responsiveness to EGFR-TRKI treatment, LUAD cells will inevitably develop acquired EGFR-TKI resistance under drug selective pressure. Secondary mutations, including EGFR-T790M mutations and other mutations within the EGFR domain, mutations in MAPK, PI3K and cell cycle genes and amplifications of other oncogenes lead to acquired drug resistance of LUAD cells to EGFR-TKI (50,51). In addition, 20–30% of LUAD patients with EGFR sensitive mutations are insensitive to EGFR-TKI and display intrinsic drug resistance (14). Therefore, it is crucial to understand the mechanisms of sensitivity and resistance to currently available therapies in this subset of lung cancers.
Previous data have shown that ferroptosis regulatory system’s inactivation plays a key role in tumor occurrence and development. Against this background, researchers have recently proposed various schemes to induce ferroptosis (52). The efficacy of ferroptosis induction therapy in treating EGFR mutant LUAD and EGFR-TKI-resistant LUAD has not been reported but it has been proven that the activation of the EGFR pathway can increase the expression of the proteins in the lipid peroxide reduction system (25-29) while also promote the accumulation of intrinsic lipid peroxides (53-55). For example, the activation of the EGFR-p62-Keap1-NRF2 pathway in tumor cells can induce xCT protein expression and contribute to ferroptosis resistance (27). In gliomas, the activation of the EGFR pathway can induce the expression of xCT and enhance the malignant potential of tumor cells (26). In the testis of Drosophila melanogaster, the activation of the EGFR pathway can support the lipid homeostasis in testicular stem cells by enhancing autophagy. However, cysteine deficiency causes EGFR mutant human breast cancer cells to be more sensitive to ROS-dependent ferroptosis (29), and the activation of EGFR-MMP3-Rac1b and EGFR-Rac1-NOX2 pathways can lead to an increase of intrinsic ROS (53-55). Therefore, it can be reasonably inferred that in LUAD cell lines which are sensitive or resistance to EGFR-TKIs, the inactivation of the lipid peroxide reduction system will irreversibly increase the level of lipid peroxide and enhance the sensitivity of cells to ferroptosis induction, as already shown in breast cancer (29).
To verify our inference, we first evaluated the efficiency of ferroptosis inducers, Erastin and RSL3, on the intrinsic and acquired EGFR-TKI resistant LUAD cells and found that ferroptosis inducers at lower than conventional doses could significantly inhibit the cell proliferation of them. These results suggested that ferroptosis induction could be used to treat intrinsic or acquired EGFR-TKI-resistant LUAD. To further dissect the therapeutic effect of ferroptosis inducers, we also screened potential drugs that could enhance tumor sensitivity to ferroptosis by combining WGCNA analysis and Connectivity Map techniques. Among all the drugs with P<0.05, the antitumor drug Vorinostat, already clinical used (56,57), had the highest specificity. By cytological experiments, we identified that Vorinostat significantly enhanced the ferroptosis induction effect of Erastin and the promoting effect of Vorinostat on ferroptosis was related to the inhibition of the expression of xCT, the key protein regulating ferroptosis.
Also, some literature showed that Vorinostat could increase the production of reactive oxidative species (ROS) (58-60), while the increase of intracellular ROS could promote lipid auto-oxidation (61) and enhance the sensitivity of tumor cells to ferroptosis. In cancer cells exposed to Erastin treatment, a compensatory upregulation of SCL7A11 can be observed (62). In our study, we found that HDAC inhibitor Vorinostat could downregulate SLC7A11, thus blocking the compensatory upregulation of SCL7A11 induced by Erastin treatment, consequently enhancing the treatment effect of Erastin.
Consistent with our findings, Zang et al. showed that Vorinostat monotherapy can significantly overcome lung cancer cells’ resistance to the first, second, and third-generation EGFR-TKI (48). However, another clinical trial showed that the addition of Vorinostat to erlotinib failed to add clinical benefit when trialed in patients with EGFR mutated advanced LUAD, previously treated with erlotinib, however, this study contained patients with T790M mutations which would not be expected to derive benefit from this combination. Its addition to osimertinib at the time of progressive disease, or to prevent intrinsic or acquired resistance, has not been studied in a clinical setting (63).
Meanwhile, our study had a number of limitations. First, our experiments were limited to in vitro scenarios and further studies should be performed in vivo. Second, we did not verify the efficacy of ferroptosis induction to overcome intrinsic and acquired EGFR-TKI resistance in LUAD patients. Third, limited panel of cell lines were analyzed. Fourth, we didn’t use EGFR wild type LUAD cell lines as control groups.
In conclusion, this study proved Vorinostat downregulated the expression of xCT in EGFR mutant LUAD cells and enhanced the effect of ferroptosis induction therapy. Therefore, the combination of Vorinostat (or another HDACi) and an established therapy that limits glutathione import or production shows a certain therapeutic prospect in treating EGFR-TKI resistant LUAD. Risk-benefit balance should be carefully considered before using this combination in patients due to the potential high toxicity of it.
Acknowledgments
The authors appreciate the academic support from AME Lung Cancer Collaborative Group.
Funding: This research was supported by the Science and Technology Development project of Shanghai Chest Hospital (to F Yao) and National Natural Science Foundation of China (NSFC82072570 to F Yao).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-21-303
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-21-303
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-21-303). Dr. Yao reports that this research was supported by the Science and Technology Development project of Shanghai Chest Hospital and National Natural Science Foundation of China (NSFC82072570). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study research did not involve human or animals, thus ethical approval was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Xu J, Zhang Y, Jin B, et al. Efficacy of EGFR tyrosine kinase inhibitors for non-adenocarcinoma lung cancer patients harboring EGFR-sensitizing mutations in China. J Cancer Res Clin Oncol 2016;142:1325-30. [Crossref] [PubMed]
- Xu J, Zhang X, Yang H, et al. Comparison of outcomes of tyrosine kinase inhibitor in first- or second-line therapy for advanced non-small-cell lung cancer patients with sensitive EGFR mutations. Oncotarget 2016;7:68442-8. [Crossref] [PubMed]
- Xu J, Jin B, Chu T, et al. EGFR tyrosine kinase inhibitor (TKI) in patients with advanced non-small cell lung cancer (NSCLC) harboring uncommon EGFR mutations: A real-world study in China. Lung Cancer 2016;96:87-92. [Crossref] [PubMed]
- Graham RP, Treece AL, Lindeman NI, et al. Worldwide Frequency of Commonly Detected EGFR Mutations. Arch Pathol Lab Med 2018;142:163-7. [Crossref] [PubMed]
- Xu J, Yang H, Jin B, et al. EGFR tyrosine kinase inhibitors versus chemotherapy as first-line therapy for non-small cell lung cancer patients with the L858R point mutation. Sci Rep 2016;6:36371. [Crossref] [PubMed]
- Ohe Y, Imamura F, Nogami N, et al. Osimertinib versus standard-of-care EGFR-TKI as first-line treatment for EGFRm advanced NSCLC: FLAURA Japanese subset. Jpn J Clin Oncol 2019;49:29-36. [Crossref] [PubMed]
- Wu YL, Cheng Y, Zhou X, et al. Dacomitinib versus gefitinib as first-line treatment for patients with EGFR-mutation-positive non-small-cell lung cancer (ARCHER 1050): a randomised, open-label, phase 3 trial. Lancet Oncol 2017;18:1454-66. [Crossref] [PubMed]
- Schuler M, Paz-Ares L, Sequist LV, et al. First-line afatinib for advanced EGFRm+ NSCLC: Analysis of long-term responders in the LUX-Lung 3, 6, and 7 trials. Lung Cancer 2019;133:10-9. [Crossref] [PubMed]
- Fukuoka M, Wu YL, Thongprasert S, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS). J Clin Oncol 2011;29:2866-74. [Crossref] [PubMed]
- Piotrowska Z, Niederst MJ, Karlovich CA, et al. Heterogeneity Underlies the Emergence of EGFR(T790) Wild-Type Clones Following Treatment of T790M-Positive Cancers with a Third-Generation EGFR Inhibitor. Cancer Discov 2015;5:713-22. [Crossref] [PubMed]
- Recondo G, Facchinetti F, Olaussen KA, et al. Making the first move in EGFR-driven or ALK-driven NSCLC: first-generation or next-generation TKI? Nat Rev Clin Oncol 2018;15:694-708. [Crossref] [PubMed]
- Tumbrink HL, Heimsoeth A, Sos ML. The next tier of EGFR resistance mutations in lung cancer. Oncogene 2021;40:1-11. [Crossref] [PubMed]
- Wang J, Wang B, Chu H, et al. Intrinsic resistance to EGFR tyrosine kinase inhibitors in advanced non-small-cell lung cancer with activating EGFR mutations. Onco Targets Ther 2016;9:3711-26. [Crossref] [PubMed]
- Bebber CM, Müller F, Prieto Clemente L, et al. Ferroptosis in Cancer Cell Biology. Cancers (Basel) 2020;12:164. [Crossref] [PubMed]
- Zhang Y, Shi J, Liu X, et al. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat Cell Biol 2018;20:1181-92. [Crossref] [PubMed]
- Wu J, Minikes AM, Gao M, et al. Intercellular interaction dictates cancer cell ferroptosis via NF2-YAP signalling. Nature 2019;572:402-6. [Crossref] [PubMed]
- Viswanathan VS, Ryan MJ, Dhruv HD, et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 2017;547:453-7. [Crossref] [PubMed]
- Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 2012;149:1060-72. [Crossref] [PubMed]
- Dixon SJ, Patel DN, Welsch M, et al. Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. Elife 2014;3:e02523 [Crossref] [PubMed]
- Yang WS. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014;156:317-31. [Crossref] [PubMed]
- Jiang L, Kon N, Li T, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 2015;520:57-62. [Crossref] [PubMed]
- Fan Z, Wirth AK, Chen D, et al. Nrf2-Keap1 pathway promotes cell proliferation and diminishes ferroptosis. Oncogenesis 2017;6:e371 [Crossref] [PubMed]
- Bersuker K, Hendricks JM, Li Z, et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 2019;575:688-92. [Crossref] [PubMed]
- Zhang MZ, Wang Y, Paueksakon P, et al. Epidermal growth factor receptor inhibition slows progression of diabetic nephropathy in association with a decrease in endoplasmic reticulum stress and an increase in autophagy. Diabetes 2014;63:2063-72. [Crossref] [PubMed]
- Tsuchihashi K, Okazaki S, Ohmura M, et al. The EGF Receptor Promotes the Malignant Potential of Glioma by Regulating Amino Acid Transport System xc Cancer Res 2016;76:2954-63. [Crossref] [PubMed]
- Sun X, Ou Z, Chen R, et al. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology 2016;63:173-84. [Crossref] [PubMed]
- Sênos Demarco R, Uyemura BS, Jones DL. EGFR Signaling Stimulates Autophagy to Regulate Stem Cell Maintenance and Lipid Homeostasis in the Drosophila Testis. Cell Rep 2020;30:1101-16.e5. [Crossref] [PubMed]
- Poursaitidis I, Wang X, Crighton T, et al. Oncogene-Selective Sensitivity to Synchronous Cell Death following Modulation of the Amino Acid Nutrient Cystine. Cell Rep 2017;18:2547-56. [Crossref] [PubMed]
- Yang H, Zhao L, Gao Y, et al. Pharmacotranscriptomic Analysis Reveals Novel Drugs and Gene Networks Regulating Ferroptosis in Cancer. Cancers (Basel) 2020;12:3273. [Crossref] [PubMed]
- Chen T, Luo J, Gu Y, et al. Comprehensive analysis of circular RNA profiling in AZD9291-resistant non-small cell lung cancer cell lines. Thorac Cancer 2019;10:930-41. [Crossref] [PubMed]
- Yang H, Hall SRR, Sun B, et al. NF2 and Canonical Hippo-YAP Pathway Define Distinct Tumor Subsets Characterized by Different Immune Deficiency and Treatment Implications in Human Pleural Mesothelioma. Cancers (Basel) 2021;13:1561. [Crossref] [PubMed]
- Wang L, Leite de Oliveira R, Huijberts S, et al. An Acquired Vulnerability of Drug-Resistant Melanoma with Therapeutic Potential. Cell 2018;173:1413-25.e14. [Crossref] [PubMed]
- Xu D, Liang SQ, Yang H, et al. Increased sensitivity to apoptosis upon endoplasmic reticulum stress-induced activation of the unfolded protein response in chemotherapy-resistant malignant pleural mesothelioma. Br J Cancer 2018;119:65-75. [Crossref] [PubMed]
- Xu D, Liang SQ, Yang H, et al. CRISPR Screening Identifies WEE1 as a Combination Target for Standard Chemotherapy in Malignant Pleural Mesothelioma. Mol Cancer Ther 2020;19:661-72. [Crossref] [PubMed]
- Xu D, Yang H, Yang Z, et al. Endoplasmic Reticulum Stress Signaling as a Therapeutic Target in Malignant Pleural Mesothelioma. Cancers (Basel) 2019;11:1502. [Crossref] [PubMed]
- Rees MG, Seashore-Ludlow B, Cheah JH, et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat Chem Biol 2016;12:109-16. [Crossref] [PubMed]
- Zhang T, Yang H, Sun B, et al. Four hub genes regulate tumor infiltration by immune cells, antitumor immunity in the tumor microenvironment, and survival outcomes in lung squamous cell carcinoma patients. Aging (Albany NY) 2021;13:3819-42. [Crossref] [PubMed]
- Yang H, Xu D, Yang Z, et al. Systematic Analysis of Aberrant Biochemical Networks and Potential Drug Vulnerabilities Induced by Tumor Suppressor Loss in Malignant Pleural Mesothelioma. Cancers (Basel) 2020;12:2310. [Crossref] [PubMed]
- Subramanian A, Narayan R, Corsello SM, et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017;171:1437-52.e17. [Crossref] [PubMed]
- Yang H, Yao F, Marti TM, et al. Beyond DNA Repair: DNA-PKcs in Tumor Metastasis, Metabolism and Immunity. Cancers (Basel) 2020;12:3389. [Crossref] [PubMed]
- Yang H, Yao F, Davis PF, et al. CD73, Tumor Plasticity and Immune Evasion in Solid Cancers. Cancers (Basel) 2021;13:177. [Crossref] [PubMed]
- Yang H, Xu D, Schmid RA, et al. Biomarker-guided targeted and immunotherapies in malignant pleural mesothelioma. Ther Adv Med Oncol 2020;12:1758835920971421 [Crossref] [PubMed]
- Choi YJ, Rho JK, Jeon BS, et al. Combined inhibition of IGFR enhances the effects of gefitinib in H1650: a lung cancer cell line with EGFR mutation and primary resistance to EGFR-TK inhibitors. Cancer Chemother Pharmacol 2010;66:381-8. [Crossref] [PubMed]
- Li X, Fu Q, Zhu Y, et al. CD147-mediated glucose metabolic regulation contributes to the predictive role of (18) F-FDG PET/CT imaging for EGFR-TKI treatment sensitivity in NSCLC. Mol Carcinog 2019;58:247-57. [Crossref] [PubMed]
- Iommelli F, De Rosa V, Gargiulo S, et al. Monitoring reversal of MET-mediated resistance to EGFR tyrosine kinase inhibitors in non-small cell lung cancer using 3'-deoxy-3'-[18F]-fluorothymidine positron emission tomography. Clin Cancer Res 2014;20:4806-15. [Crossref] [PubMed]
- Suda K, Tomizawa K, Fujii M, et al. Epithelial to mesenchymal transition in an epidermal growth factor receptor-mutant lung cancer cell line with acquired resistance to erlotinib. J Thorac Oncol 2011;6:1152-61. [Crossref] [PubMed]
- Zang H, Qian G, Zong D, et al. Overcoming acquired resistance of epidermal growth factor receptor-mutant non-small cell lung cancer cells to osimertinib by combining osimertinib with the histone deacetylase inhibitor panobinostat (LBH589). Cancer 2020;126:2024-33. [Crossref] [PubMed]
- Ettinger DS, Wood DE, Aisner DL, et al. Non-Small Cell Lung Cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2017;15:504-35. [Crossref] [PubMed]
- Kashima K, Kawauchi H, Tanimura H, et al. CH7233163 overcomes osimertinib resistant EGFR-Del19/T790M/C797S mutation. Mol Cancer Ther 2020; [Crossref] [PubMed]
- Leonetti A, Sharma S, Minari R, et al. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer 2019;121:725-37. [Crossref] [PubMed]
- Tarangelo A, Dixon SJ. Nanomedicine: An iron age for cancer therapy. Nat Nanotechnol 2016;11:921-2. [Crossref] [PubMed]
- Jin X, Jin X, Sohn YW, et al. Blockade of EGFR signaling promotes glioma stem-like cell invasiveness by abolishing ID3-mediated inhibition of p27(KIP1) and MMP3 expression. Cancer Lett 2013;328:235-42. [Crossref] [PubMed]
- Radisky DC, Levy DD, Littlepage LE, et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature 2005;436:123-7. [Crossref] [PubMed]
- Sheng ZG, Huang W, Liu YX, et al. Ofloxacin induces apoptosis via β1 integrin-EGFR-Rac1-Nox2 pathway in microencapsulated chondrocytes. Toxicol Appl Pharmacol 2013;267:74-87. [Crossref] [PubMed]
- Gray JE, Saltos A, Tanvetyanon T, et al. Phase I/Ib Study of Pembrolizumab Plus Vorinostat in Advanced/Metastatic Non-Small Cell Lung Cancer. Clin Cancer Res 2019;25:6623-32. [Crossref] [PubMed]
- Takeuchi S, Hase T, Shimizu S, et al. Phase I study of vorinostat with gefitinib in BIM deletion polymorphism/epidermal growth factor receptor mutation double-positive lung cancer. Cancer Sci 2020;111:561-70. [Crossref] [PubMed]
- Carew JS, Giles FJ, Nawrocki ST. Histone deacetylase inhibitors: mechanisms of cell death and promise in combination cancer therapy. Cancer Lett 2008;269:7-17. [Crossref] [PubMed]
- Mariadason JM. HDACs and HDAC inhibitors in colon cancer. Epigenetics 2008;3:28-37. [Crossref] [PubMed]
- Marks PA, Xu WS. Histone Deacetylase Inhibitors: Potential in Cancer Therapy. J Cell Biochem 2009;107:600-8. [Crossref] [PubMed]
- Miyamoto K, Watanabe M, Boku S, et al. xCT Inhibition Increases Sensitivity to Vorinostat in a ROS-Dependent Manner. Cancers (Basel) 2020;12:827. [Crossref] [PubMed]
- Zhang Y, Koppula P, Gan B. Regulation of H2A ubiquitination and SLC7A11 expression by BAP1 and PRC1. Cell Cycle 2019;18:773-83. [Crossref] [PubMed]
- Reguart N, Rosell R, Cardenal F, et al. Phase I/II trial of vorinostat (SAHA) and erlotinib for non-small cell lung cancer (NSCLC) patients with epidermal growth factor receptor (EGFR) mutations after erlotinib progression. Lung Cancer 2014;84:161-7. [Crossref] [PubMed]


