
Diagnostic and prognostic biomarkers in oligometastatic non-small cell lung cancer: a literature review
Introduction
Since the first observations on growth and the metastatic spreading of non-small cell lung cancer (NSCLC), it has been clear that some tumors remained confined and indolent in few organs for a long time (1). The “oligometastatic” condition—hereafter called oligometastatic NSCLC—was initially defined as an intermediate stage between locally advanced and widely disseminated disease (1). However, this definition was inaccurate and did not discriminate between primary oligometastatic disease and oligorecurrence, and different cut-off numbers of metastases or organs involved were used (1).
Recently, the European Organization for Research and Treatment of Cancer Lung Cancer Group (EORTC-LCG) published a consensus about the clinical definition of oligometastatic NSCLC (2) and described different patterns among de-novo status, repeat or induced oligometastatic disease in collaboration with the European Society of Radiotherapy (ESTRO) (3).
All definitions summarize a phenotypic rather than a genotypic condition; however, describing genetic features and microRNA signatures may be pivotal in the diagnostic process of oligometastatic disease (4). In clinical practice, genotyping to identify oligometastatic-NSCLC (OM-NSCLC) is not feasible, and the diagnosis of a clinical oligometastatic status per se is a prognostic factor.
Indeed, among clinical factors, metachronous versus synchronous metastases, N-stage, and adenocarcinoma histology may stratify the risk of progressive disease and death (5).
Lacking biomarkers of OM-NSCLC, it is impossible to differentiate an oligometastatic disease that grows slowly and remains in this clinical status from an oligometastatic disease that continues to proliferate and spreads in multiple organs. Furthermore, it is not possible to distinguish the definition of metastasis in multinodular lung disease from multiple primary lung tumors with the prognostic consequences and the therapeutic strategy of the case only from histopathological evidence (6,7).
Many efforts had been done to discriminate multiple primary lung cancers from intrapulmonary metastasis. The first attempt was based on the expression of four cancer-related proteins—p53, p16, p27, and C-erbB2 (8). Subsequently, the TRACERx program, using the whole-exon sequencing, revealed a more complex genetic scenario (9). Tissue genetic profiling and liquid biopsies resulted as effective techniques to achieve the correct diagnosis of oligometastatic disease and ameliorate the individualized therapeutic strategy.
Tissue-based biomolecular biomarkers can more accurately identify those patients who might benefit from local therapy, describing mRNA expression, microRNA expression, DNA mutations, epigenetic changes. In these procedures, poor quality of tissue and tumor heterogeneity—spatial and temporal—may be limiting (10).
Blood-based biomarkers are attractive, since they do not require invasive biopsies and may explore many tumoral components such as proteins, microRNAs, circulating tumor cells, ctDNA, and exosomes. These techniques can be repeated, if necessary, and better reflect tumor molecular heterogeneity, either temporal or spatial, than a single biopsy (11,12).
ctDNA is quantitively related to tumor burden and is a more objective measure of total body disease burden than imaging. Based on these observations, ctDNA profile can be useful to determine the minimal residual disease (MRD), defined as a small volume of tumor cells remaining after treatment in patients who do not have clinical evidence of disease, in early-stage or oligometastatic solid tumors and may allow predicting the outcomes in oligometastatic patients (13). In OM-NSCLC, ctDNA profile can be applied both to identify those patients who are eligible for local ablative treatment and to follow them after ablative treatments that completely eradicate metastatic deposits and lead to a definitive cure.
This review summarizes current approaches of diagnosis, monitoring, and prognosis of OM-NSCLC and describes the fast-evolving therapeutic scenario of this disease. We present the following article in accordance with the Narrative Review reporting checklist (available at https://dx.doi.org/10.21037/tlcr-20-1067).
Methods
We performed an updated literature search for papers published up to October 2020 about the role of diagnostic and prognostic biomarkers for OM-NSCLC throughout different medical research databases like PubMed, Scopus, and Web of Science, as well as an evaluation of abstracts reported on principal international cancer congresses (ASCO, ESMO, IASLC) websites.
We employed for the search the following terms: “oligometastatic non-small cell lung cancer”, “biomolecular markers”, “predictive biomarkers”, “prognostic biomarkers”, “liquid biopsy”, and “oligometastatic non-small cell lung cancer”. We retrieved and analyzed also data of completed or ongoing clinical trials about this specific topic.
Given the hugeness of the data concerning the predictive and prognostic factors of response to treatments, we have focused on the technological advancement of circulant biomolecular factors in OM NSCLC pathology, starting from apparently simpler models such as oncogene addicted tumors to evaluate themselves in the light of treatment with immune-checkpoint inhibitors.
Biological techniques
Interventional pathologists play a pivotal role in the molecular characterization of OM-NSCLC, triaging the appropriate technique for the mini-invasive evaluation of the dynamic changes related to the genomic landscape of tumors. Cytological specimens may be the first ideal source of DNA for next-generation sequencing (NGS); fine needle aspirations (FNAs) are safe and cost-effective procedures that allow an optimal sampling of deep lesions via radiological guides (ultrasound/computed tomography). Traditional or liquid-based preparations are suitable for the application of exome sequencing or customized panels and, especially, May Grunwald Giemsa (MMG)-stained slides can be easily microdissected to obtain enriched samples. Supernatants may provide adequate material for the detection of driver mutations in oncogene-addicted cancers, helping to avoid the sacrifice of diagnostic stained smears (14). Especially during the natural history of long-lasting cancers, the possible contribution of core-needles as repeat biopsies is debated. Traditionally, processed formalin-fixed paraffin-embedded (FFPE) samples have been compared to paired PAXgene® tissue fixed paraffin-embedded (PFPE) tissues, which might be superior for DNA and RNA integrity, particularly in low-yield samples (15). Cytology and micro-histological approaches might work in a complementary fashion (16), however, when neoplastic sites are not accessible or clinicians have not the possibility to apply an interventional procedure with the requested advantage, the recourse to liquid biopsy is completely justified. The best way to guarantee multitasking management of precious oncological samples is the careful choice of the most appropriate way to take on the bioptic target and the medium to maximize the extraction of nucleic acids from small specimens.
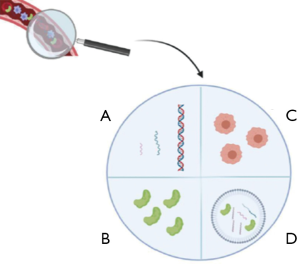
The term “liquid biopsy” defines not only cell-free DNA (cfDNA), shed into the bloodstream by tumor and non-tumor cells, but also other relevant biological molecules and macrostructures, such as microRNA, circulating free RNA, extracellular vesicles, and circulating tumor cells (Figure 1) (17). CtDNA represents the specific fraction of tumor-derived cfDNA to assess the molecular status of epidermal growth factor receptor (EGFR) (18-21) at the resistance from first-line treatment with a first or second-generation EGFR tyrosine kinase inhibitors (TKIs) or the basal setting when tumor tissue is absent or not adequate for predictive analysis (18).
At the moment, the correlation between ctDNA levels and tumor burden (22) or the possibility to adopt liquid biopsy in non-advanced stage settings [such as cancer interception, early detection, and MRD (23,24)], represent research hot-topics.
A correlation among ctDNA levels and disease burden and specific metastatic sistes has been widely demonstrated (22). In patients with advanced-stage solid, the ctDNA concentration in the bloodstream is about 100 times higher concerning early-stage patients (25). In a recent prospective case-control sub-study by Liu et al, targeted methylation analysis of cfDNA was performed to detect and localize multiple cancer types across all stages on 2,482 cancer patients (>50 different cancer types) and 4207 non-cancer people (26). The overall specificity of 99.3% in cancer detection was reached. Interestingly, the authors highlighted that the detection rate was higher in advanced stages. As far as tumors are concerned, sensitivity was 18% in stage I, 43% in stage II, 81% in stage III, and 93% in stage IV (26).
In this scenario, another key factor is represented by the tumor dimension. An overall tumor volume of 10 cm3 seems to be the cut-off value to reach an adequate sensitivity level when considering ctDNA analysis (27). More recently, as discussed below, the integration of different biological levels (genetics, transcriptomics, and proteomics) may be a fascinating tool to overcome the limitations of ctDNA analysis, to implement liquid biopsy even for early-stage patients and cancer interception (28).
Data from proteomic analyses indicated that proteins of extracellular vesicles and particles of 426 human samples could serve as reliable biomarkers for cancer detection (29).
Another crucial issue related to the ctDNA analysis in solid tumors is associated with the specific metastatic site. The ctDNA “shareability” into the bloodstream is strictly dependent, not only on the tumor type (Table 1) (30-35) but also on the metastatic site. As an example, in a really interesting pooled analysis by Passiglia et al, considering a set of ten studies, for a total of 1425 NSCLC patients, the sensitivity of ctDNA based EGFR mutation testing was significantly higher in patients with M1b vs. M1a disease stage (odds ratio, OR: 5.09; 95% CI: 2.93–8.84) regardless the use of digital (OR: 5.85, 95% CI: 3.56–9.60) or non-digital polymerase chain reaction technologies (OR: 2.96, 95% CI: 2.24–3.91) (36). These well-structured analyses showed that the specific metastatic site significantly affected the accuracy of ctDNA based EGFR mutations analysis in NSCLC patients (36).

Full table
In most cases, advanced-stage cancer patients show a central nervous system, pleural, or peritoneal metastasis. In a recent prospective study by Villatoro et al, 42 cerebrospinal fluid (CSF), pleural effusion, and/or ascites were used as a source of ctDNA in advanced NSCLC and melanoma patients and compared with results obtained from paired blood samples in 22 cases. The results obtained underline that fluids close to metastatic sites are superior to blood for the detection of relevant mutations (37). These important results can be useful in the oligometastatic patients, for the ctDNA molecular characterization to define the treatment strategy.
Also in this scenario, as previously discussed for tumor stage and size, the integration of different “omics” levels, may represent a key “weapon” to overcome the limitations related to the metastatic site location, not only for NSCLC patients but also for other types of solid tumors.
Multidimensional liquid biopsy analysis
The integration between the epigenetic fingerprints and the mutational profile of ctDNA is exemplified by the GRAIL Inc. program (26). Based on the evidence of Liu et al. (26), this innovative approach is focused on ctDNA sequence methylation assessment, through the analysis of 100,000 methylation regions (covering ~1 million CpG sites), by using a highly efficient targeted bisulfite NGS and machine learning, able to identify the abnormal methylated ctDNA region.
Another example of different omics level integration is represented by CancerSEEK (Thrive Earlier Detection Corp.) that combines the detection of ctDNA mutation profile and protein expression associated with eight types of cancers, including ovarian, liver, stomach, pancreatic, and esophageal cancers, with a median sensitivity of 70%.
Despite the exciting results, both GRAIL and CancerSEEK (and other similar approaches) are not ready for clinical implementation and larger studies are currently ongoing for the clinical validation of these novel tools.
Liquid biopsy real-time monitoring for OM-NSCLC
One of the most fascinating opportunities offered by liquid biopsies is the possibility to track clonal evolution during anticancer treatments, pinpointing the emergence of resistant clones before radiographic progression and monitoring MRD. The term MRD refers to the evidence of a small number of cancer cells that remain in the body during or after treatment and are associated with prognostic and therapeutic relevance. Liquid biopsy is particularly suitable for this scope due to its minimally invasive nature that allows multiple evaluations over time without significant risks for the patient.
Several recent studies have evaluated the use of cfDNA analysis for real-time monitoring during EGFR TKIs in EGFR-mutated NSCLCs using different methodologies, including digital droplet PCR (ddPCR) (38-41), next-generation sequencing (NGS) (37,40), and Peptide nucleic acid–zip nucleic acid clamp PCR (32). Collectively, these studies have shown a promising role for longitudinal monitoring of EGFR mutations in cfDNA through liquid biopsy, allowing the identification of acquired resistance mechanisms, such as the secondary EGFR mutation T790M after the 1st/2nd generation EGFR TKIs, even before radiographic progression. The use of NGS platforms offers the opportunity to simultaneously evaluate different mechanisms of resistance with high sensitivity and should be preferred over PCR-based methods (42). Whether the identification of mechanisms of acquired resistance in cfDNA before radiographic progression should lead to a therapeutic switch from a line of therapy to another (for instance, different EGFR TKIs) is a matter of debate and is currently under evaluation in the EORTC phase II study APPLE trial (NCT02856893).
In the context of OM-NSCLC and oligoprogressive disease, the use of cfDNA analysis could be of high clinical relevance (Figure 2), since multiple clinical studies have recently reported that cfDNA dynamics are a strong prognostic factor during anticancer therapies in advanced NSCLC, including EGFR TKIs (43-45) and immune checkpoint blockage with PD-(L)1 inhibitors (46-48). The dynamics of ctDNA have been shown to correlate with the outcome of advanced NSCLC patients treated with PD-(L)1 inhibitors and could better discriminate equivocal radiographic patterns of response, such as pseudoprogression and/or oligoprogression. In addition, ctDNA changes seem to correlate with pathologic response in early NSCLC after neoadjuvant PD-(L)1 blockage (46). Using an ultrasensitive liquid biopsy test (Cancer Personalized Profiling By Deep Sequencing, CAPP-Seq), Zhang et al. showed that ctDNA analysis can noninvasively identify MRD in NSCLC patients with long-term benefit to PD-(L)1 blockade (≥12 months), with 93% of patients with undetectable ctDNA levels progression-free versus none of the patients with detectable ctDNA (P<0.0001) (49). Albeit limited by small sample size (n=31), the study is hypothesis-generating and the use of liquid biopsy might allow personalized strategies for a treatment duration of immune checkpoint blockade and enable early intervention in patients at high risk for progression (49).
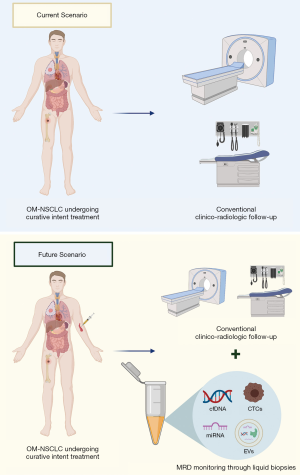
Data on OM-NSCLC are scant. Recently, an exploratory analysis of a randomized phase II study evaluating local consolidative therapy versus maintenance therapy or observation was reported (50). Plasma NGS was performed on 21 patients using a 1,021 cancer gene panel. At early follow-up timepoints, patients treated with local consolidative therapy were associated with a lower detected mutation burden. Furthermore, in a small subset of patients (n=6) with available serial samples for ctDNA analysis, the first increase of ctDNA mutation burden preceded radiographic progression by a median of 6.7 months (range, 2.9–17.9 months) (49). These results are in line with previous findings in early-stage NSCLC (stage I–III), where ctDNA detection after a curative intent treatment precedes radiographic progression in 72% of the patients by a median of 5.2 months (13). Furthermore, recently Chabon et al. showed that pretreatment ctDNA levels in stage I-IIIA NSCLC were associated with prognosis in terms of both disease-free and distant metastasis-free survival (51), suggesting that persistence of ctDNA detection after curative-intent treatments might reflect the presence of micrometastases (52). Collectively, these results indicate that ctDNA levels are associated with tumor burden and might be used as a minimally invasive monitoring tool after curative intent therapies in early-stage NSCLC and/or local consolidative therapies in OM-NSCLC, allowing personalized therapeutic strategies in positive cases.
Whether ctDNA changes during treatment of OM-NSCLC could represent a novel reliable biomarker will be further investigated in the ongoing phase II EXTEND basket trial (NCT03599765) (53), assessing the efficacy and safety of upfront local consolidative therapy in OM solid tumors, and the phase III LONESTAR trial (NCT03391869) (54), evaluating the impact of local consolidative therapy after nivolumab-ipilimumab in stage IV NSCLC.
Prognostic and predictive biomarkers for the OM-NSCLC
Two clinical factors have been associated with the OS and prognostic stratification of patients with OM-NSLC, i.e., the type of metastatic presentation (synchronous versus metachronous) and N status (5), and several candidate biomarkers are under investigation to predict the outcome of patients with OM-NSCLC and their response to treatments.
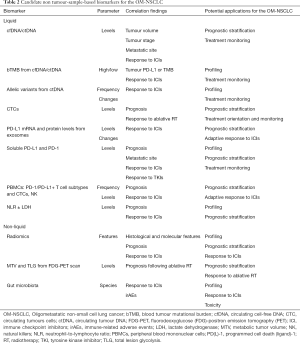
Besides tumor-sample-based biomarkers, such as the PD-L1 expression, the high-microsatellite instability (MSI-H), the tumor mutational burden (TMB), the presence of tumor-infiltrating lymphocytes (TILs), and gene expression profiling (GEP) (55,56) liquid and other non-tumor sample-based biomarkers may offer the advantage to overcome the lack of tissue, the tumor heterogeneity, and the different adaptive mechanisms of tumors cells to treatments (57) and will be briefly described (Table 2).
Full table
Relevant vectors from liquid biopsies are cfDNA/ctDNA (and the associated level of blood TMB), peripheral blood mononuclear cells (PBMSCs), soluble mediators (i.e., proteins), circulating tumor cells (CTCs), exosomes, and microRNA (58).
As previously discussed in this review, in EGFR mutated NSCLCs, the presence of ctDNA and its quantity can be related to tumor volume, stage, and possible discrimination between specific metastatic sites (i.e., M1b vs. M1a) (22,25,35). Their sensitivity could be variable depending on tumor shedding. The presence and the levels of cfDNA/ctDNA could be explored in the OM-NSCLC for prognostic stratification and treatment monitoring.
Based on cfDNA and ctDNA, their levels, blood TMB (bTMB), and allelic variant frequencies have been explored in advanced NSCLC as biomarkers predictive of PD-1/PD-L1 tumor expression and outcomes under immune-checkpoint inhibitors (ICIs). A high bTMB has been correlated with the favorable clinical outcome under ICIs independently by PD-L1 expression on tumor tissue (59) and could be correlated to tissue TMB (60). Changes in levels of ctDNA (61) and the allelic frequency of distinct mutations measured in ctDNA (47,61,62) showed concordance with disease response to ICIs and could assist radiographic assessments (58). Therefore, these biomarkers could be helpful to better define genomic profiling between the OM-NSCLCs and towards the non-OM-NSCLC and to monitor treatment response to treatment.
Other types of circulating biomarkers
As previously discussed, the emerging concept of liquid biopsy includes also other biological relevant molecules and macrostructures, such as miRNA, cell-free RNA, extracellular vesicles and circulating tumor cells (16).The simultaneous analysis of these different analytes, in combination with data obtained from the ctDNA mutational analysis may represent a novel advice to overcome the limitations related to the low sensitivity of liquid biopsy in some settings, such as cancer interception, early stage disease analysis and MRD (16).
The role of other liquid biopsy components, such as circulating tumor cells (CTCs) and extracellular vehicles (EVs) is far less defined in the context of OM-NSCLC. The presence of ≥5 CTCs before chemotherapy was significantly associated with worse prognosis in advanced NSCLC (52). Their role in OM-NSCLC is still unclear, albeit some indirect evidence suggests a promising role in this peculiar clinical scenario. For instance, in early-stage NSCLC (stage I) treated with stereotactic body radiotherapy (SBRT), pretreatment levels of CTCs (using a cut-off of ≥5 CTCs/mL) and posttreatment persistence are significantly associated with increased risk of recurrence outside the targeted treatment site (63). Furthermore, longitudinal CTC monitoring in patients with locally advanced NSCLC treated with chemo-radiotherapy predicted the outcome, with most of the patients having undetectable CTCs on initial post-radiotherapy draw and CTCs re-emergence predicted disease progression before radiographic evidence of recurrence (64). These results suggest a potential application of CTCs in the context of OM disease and deserve further confirmation in prospective studies.
The presence and the amount of blood CTCs could represent another potential biomarker to differentiate OM-NSCLCs and compare to non-OM-NSCLCs, to predict prognosis and inform treatment strategies. For instance, fewer CTCs were detected among NSCLC patients with OM than in those with non-OM brain metastases, while the presence of CTCs and their thresholds of ≥2 and ≥5 CTCs/7.5 mL of blood were independent factors for overall survival in the OM disease (65). This could suggest more intensive treatments in patients with OM-NSCLC with brain metastases and high levels of CTCs. Furthermore, PD-L1+ CTCs were associated with poor OS and treatment failure (58).
PD-L1 mRNA and protein levels in circulating extracellular vesicles (often referred to as exosomes) may also have the potential to predict tumor response to anti-PD1/PD-L1 antibodies. Cancer cells release exosomes from the endocytic compartment into the plasma and they play a role in influencing processes involved in tumor progression, including immunoediting and drug resistance, by transporting nucleic acids, proteins and lipids to nearby or distant cells (66). PD-L1 protein levels in circulating exosomes and changes in plasma levels of PD-L1 positive exosomes, by the analysis of PD-L1 mRNA, were related with treatment outcome to PD-1/PD-L1 inhibitors in melanoma and NSCLC patients (67,68). The presence of PD-L1 on tumor-derived exosomes may allow them to target PD-1+ CD8 T-cells as a mechanism of the adaptive response of the tumor cells to T cell reinvigoration, thus allowing them to evade immune response at the effector stage (67).
The soluble form of PD-L1 (sPD-L1) levels have not been related to the tumor PD-L1 (69-71). However, high baseline sPD-L1 levels were associated with poor prognosis and abdominal metastases (69,72), whilst low baseline sPD-L1 levels were linked to treatment efficacy (68) and increasing levels to treatment failure (70). High sPD-1 levels predicted a favorable outcome to the TKIs (73).
PBMCs include T-cells (≈70%), B-cells (≈15%), monocytes (≈5%) and natural killer cells (≈10%) whose proportion may considerably differ by showing the activity of the immune system and play an important role as immunological biomarkers (74). Furthermore, PD-1 expression can be measured on PBMCs and is elevated among CD4+ T-cells from advanced NSCLC patients compared with healthy donors, while no correlation between PD-L1 expression on tumor cells and PD-1 expression on CD4+ and CD8+ T cells was found (75). High PD-L1 CD4+ T cells were associated with unfavorable outcome in OS and PFS and failure to ICIs (75), whilst an early (within 4 weeks) increase in PD-1+ CD8+ T cells predicted better response to anti-PD1 agents (76). A high baseline number of NK-cells and their increase during ICIs was associated with treatment response (77).
The neutrophil-to-lymphocyte ratio (NLR) is a surrogate for tumor-associated inflammation and likely represents the frequency and activity of myeloid-derived suppressor cells, that hinder T-cell proliferation and expansion (78). A high NLR and its combination with high lactate dehydrogenase or tumor PD-L1 expression level were related with worse outcomes to ICIs, but not to chemotherapy (79-81).
PD-L1 protein levels in exosomes, sPD-L1/PD-1 levels, PD-1/PD-L1 expression on T cells and CTCs and NLR could, therefore, be prognostic and useful to explore possible differences in the adaptive response to PD-1/PD-L1 inhibitors in the OM-NSCLC.
However, although promising, the use in clinical practice of these soluble biomarkers is currently limited by data heterogeneity, lack of direct comparison of assays, vectors, and validations (58). No liquid biomarker has been currently tested in randomized phase III trials, although the first trial is ongoing (82).
Recently, deep learning approaches applied to the quantitative analysis of radiological images (namely the radiomics approach) has been explored as a diagnostic and risk-layering tool to characterize specific tumors features (such as the PD-L1 expression or the TMB) and predict patients’ prognosis and response to treatments by offering interpretable artificial intelligence (AI) models able to merge a high amount of information from different vectors (83). Of course, the utility of radiomics in the OM-NSCLC could vary from assisting the histological and molecular diagnosis of the tumor to profiling better this disease subgroup and predict prognosis and response to treatments.
Another imaging biomarker that could have a role for the prediction of outcome following ablative radiotherapy of the OM-NSCLC could be the metabolic burden of the disease as defined by the metabolic tumor volume and total lesion glycolysis of all lesions by the fluorodeoxyglucose positron emission tomography as both resulted as independent prognostic factors for the OS (84).
Finally, the study of gut microbiota, that could be obtained by stool cultures or molecular techniques (i.e., sequencing, metagenomics), has associated various species to the disease response and immune-related adverse events (irAEs) from ICIs and could be useful to profile the OM-NSCLC and predict disease response and toxicity from the ICIs.
Diagnostic and prognostic biomarker in oligometastatic NSCLC: are we ready from prime time?
Liquid biopsy and ctDNA profiling are usually employed in clinical practice to detect the EGFR T790M mutation and the subsequent sensitivity to Osimertinib (85). These techniques are extremely sensitive and specific and are currently recommended for T790M testing when tissue biopsy is not feasible (86). In some oligometastatic diseases, the difficulties to obtain a fresh biopsy make the liquid biopsy the unique way to employ a treatment driven by an actionable target, and different sources of ctDNA, such as liquor, have been explored. Even in these cases, Osimertinib was active in presence of EGFR T790M (87).
Dynamic changes of EGFR mutations in plasma or cerebrospinal fluid during the EGFR tyrosine kinase inhibitor treatment are directly linked to clinical activity and monitoring these fluctuations may be useful to decide the strategy in the management of intra and extracranial disease (88).
The open-label, single-arm, prospective APOLLO study enrolled patients with EGFR T790M positive NSCLC with brain metastatic spread, treated with Osimertinib (89). In twelve patients with paired cerebrospinal fluid and plasma sample, a low concordance between cerebrospinal fluid/plasma EGFR T790M status positivity (8.3%) was detected, underlying the differences of the microenvironment of intra and extracranial sites. However, a higher concordance was seen in EGFR mutation-sensitive del19 and L858R between plasma and cerebrospinal fluid (100% and 75%, respectively).
The clearance of T790M in cerebrospinal fluid after 6 weeks of treatment displays a higher intracranial response rate and a trend in better median progression-free survival.
This trial is the last example of a series of trials (90) that demonstrated that early clearance of EGFR mutation in plasma may correlate with positive activity and efficacy outcomes. The presence of EGFR T790M variant allelic frequency (VAF) rather than its clearance is predictive of Osimertinib activity (85).
The cut-off points of plasma VAF at the beginning of therapy and the convenient time to detect VAF clearance during treatment (i.e., 3–6 weeks) are still a matter of debate and the detection of resistance mutation is not enough to change accordingly the therapeutic strategy.
Even if some experiences demonstrated that the occurrence of plasma mutation resistance could anticipate the clinical progression of a median of 3 months, no prospective trials with complete accrual indicated if it is opportune to change the therapeutic behavior (91).
The ongoing APPLE trial is a unique example of a prospective clinical trial in which the therapeutic strategy is adapted after the detection of T790M mutation at fixed timepoints in ctDNA in EGFR mutant TKI treatment-naïve patients. The trial compared continuing gefitinib until disease progression according to RECIST or switching to Osimertinib when plasma T790M arose and clarified the predictive power of liquid biopsy in this setting (82).
In other oncogene-addicted lung cancers, such as ALK-driven tumors, ctDNA by calculating maximum allelic frequency correlates with tumor burden and predicts the response to ALK inhibitors. In these experiences, maximum plasma allelic frequency and ALK alteration VAF are independent surrogates of ctDNA and high disease burden.
ALK fusion/ mutations have been found in more than 2/3 of plasma analyzed patients; therefore, a personalized therapeutic strategy that chooses active ALK inhibitors according to ALK-driven resistance mutation detected would become reasonable. However, also for ALK fusion or mutated NSCLCs, no prospective trials are still published about therapeutic change driven by dynamically testing of plasma ALK VAF.
Like ALK-harboring tumors, liver, adrenal glands, and bone are metastatic sites that correlate with a higher burden of ctDNA shedding (92); for instance, the plasma assay is more sensitive when the central nervous system is the prevalent site of metastasis (93,94).
In oncogene-addicted OM-NSCLC, ctDNA profiling is an attractive tool in both diagnostic settings when the tissue biopsy is hard to collect and monitoring of therapeutic activity. However, no conclusion about the prognosis could be done without a link to an elevated ctDNA burden at the diagnosis (Figure 3) (95).
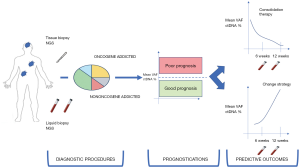
The need for coupling prognostic information and predictive activity of therapeutic strategy is more evident in the field of non-oncogene addicted oligometastatic NSCLC, eventually eligible for immunotherapy. Recently, a comprehensive analysis of ctDNA VAF from solid tumors with a large representation of lung tumors treated with durvalumab combined or not with tremelimumab demonstrated that a higher basal VAF was associated with poor prognosis while the reduction of ctDNA during treatment had a predictive meaning (49). Baseline mean or maximum VAF was associated with median OS and not with the overall response rate (ORR), tumor burden or other prognostic information, as showed in multivariate analysis (95). Therefore, ctDNA can be considered as a prognostic factor rather than a predictor of response to immune-checkpoint inhibitors.
The ctDNA dynamics correlated to the clinical benefit of immunotherapy, as the reduction of ctDNA mean VAF measured at 6 weeks was associated with ORR and OS (62).
Data that correlate mean VAF and prognosis associated with immunotherapy have been already reported, but the cut-offs of mean ctDNA VAF to distinguish poor from good prognosis have not been established yet (96).
The detection of stable or increased levels in circulating DNA VAF is an interesting phenomenon to evaluate the activity of disease when instrumental imaging scans restage a stable disease, as per RECIST criteria. Again, the lack of results from well-conducted prospective trials does not allow to draw a firm indication about the impact on therapeutic strategy.
Thus, the liquid biopsy approach may represent a valuable option in OM-NSCLC to overcome issues related to invasive tissue sampling procedures. In addition, the selection of a “liquid biopsy” more close to the metastatic site (e.g. CSF in brain metastasis) may overcome the limitations of a low shedding of tumor-related analytes into the bloodstream, leading to false-negative molecular results.
Conclusions
Clinical data available to date are not focused on OM-NSCLC setting and the scenario that is more likely to be employed arises from oncogene-addicted tumors and NSCLC during a deep response to the drug.
In the setting of NSCLC, liquid biopsy developed in early-stage disease or when a MRD is expected during active treatment is feasible; however, since a low ctDNA shedding is expected due to a low tumor burden, ultra-sensitive assays are necessary. In addition, the liquid biopsy approach can overcome the limitation of tissue-based approaches related to the spatial and temporal molecular heterogeneity that may significantly limit the adoption of the best treatment option in OM-NSCLC patients.
In OM-NSCLC, liquid biopsy can be a diagnostic tool, seeking oncogene mutation typically expressed in lung cancers; this technique may overcome the difficulties due to small tissue biopsy in low metastatic spreading disease or localized in difficult to reach the site (i.e., brain localization).
From a prognostic point of view, the mean ctDNA VAF seems to be the only biological factor that emerges from literature, even if it has not been specifically studied in the OM-NSCLC setting.
The main point is that the oligometastatic disease is currently described using clinical and morphological definitions rather than its biological features; data from prospective clinical trials in an early stage of disease, coupled with knowledge of genetic characteristic of lung tumors are warranted since they may further clarify diagnostic and prognostic features of this sub-group of disease (97). These efforts would lead to improving the possibility to eradicate the residual disease in these low burden tumoral settings, enhancing the definitive cure perspectives.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Maurizio Infante & Thierry Berghmans) for the series “Oligometastatic NSCLC: definition and treatment opportunities” published in Translational Lung Cancer Research. The article has undergone external peer review.
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://dx.doi.org/10.21037/tlcr-20-1067
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tlcr-20-1067). The series “Oligometastatic NSCLC: definition and treatment opportunities” was commissioned by the editorial office without any funding or sponsorship. DC has received personal fees (as speaker bureau and/or advisor) from MSD, BMS, Astra Zeneca, Roche, Eli Lilly, Amgen, and Novartis, unrelated to the current work. UM has received personal fees (as consultant and/or speaker bureau) from Boehringer Ingelheim, Roche, MSD, Amgen, Thermo Fisher Scientifics, Eli Lilly, Diaceutics, GSK, Merck, and AstraZeneca, unrelated to the current work. PF has received personal fees (as consultant and/or speaker bureau) from AstraZeneca, Roche, MSD, Amgen and Merck, unrelated to the current work. RA has received personal fees from Astra Zeneca, MSD and Novartis, unrelated to the current work. BGL reports personal fees from Janssen-Cilag, Boehringer Ingelheim, Roche and AstraZeneca/MedImmune, non-financial support from Bristol-Myers Squibb, Pierre Fabre and Ipsen, outside the submitted work. RC reports grants from Lung Cancer Research Foundation-Pfizer Grant 2019, payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events from MSD, Astra Zeneca, Roche, participation on a Data Safety Monitoring Board or Advisory Board from Inivata, ArcherDx, MD Serono, BMS, Novartis, Boston Pharmaceuticals, and other financial or non-financial interests: research collaboration from GuardantHealth, IALSC Vice President: ISLB (International Society of Liquid Biopsy), Educational Chair: OLA Oncology Latin American Association - Faculty for ASCO International Scientific Committee Member at ESO (European School of Oncology). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hellman S, Weichselbaum RR. Oligometastases. J Clin Oncol 1995;13:8-10. [Crossref] [PubMed]
- Levy A, Hendriks LEL, Berghmans T, et al. EORTC Lung Cancer Group survey on the definition of NSCLC synchronous oligometastatic disease. Eur J Cancer 2019;122:109-14. [Crossref] [PubMed]
- Guckenberger M, Lievens Y, Bouma AB, et al. Characterisation and classification of oligometastatic disease: a European Society for Radiotherapy and Oncology and European Organisation for Research and Treatment of Cancer consensus recommendation. Lancet Oncol 2020;21:e18-28. [Crossref] [PubMed]
- Uppal A, Wightman SC, Mallon S, et al. 14q32-encoded microRNAs mediate an oligometastatic phenotype. Oncotarget 2015;6:3540-52. [Crossref] [PubMed]
- Ashworth AB, Senan S, Palma DA, et al. An individual patient data metaanalysis of outcomes and prognostic factors after treatment of oligometastatic non-small-cell lung cancer. Clin Lung Cancer 2014;15:346-55. [Crossref] [PubMed]
- Girard N, Ostrovnaya I, Lau C, et al. Genomic and mutational profiling to assess clonal relationships between multiple non-small cell lung cancers. Clin Cancer Res 2009;15:5184-90. [Crossref] [PubMed]
- Girard N, Deshpande C, Lau C, et al. Comprehensive histologic assessment helps to differentiate multiple lung primary nonsmall cell carcinomas from metastases. Am J Surg Pathol 2009;33:1752-64. [Crossref] [PubMed]
- Ono K, Sugio K, Uramoto H, et al. Discrimination of multiple primary lung cancers from intrapulmonary metastasis based on the expression of four cancer-related proteins. Cancer 2009;115:3489-500. [Crossref] [PubMed]
- Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2109-21. [Crossref] [PubMed]
- Pitroda SP, Khodarev NN, Huang L, et al. Integrated molecular subtyping defines a curable oligometastatic state in colorectal liver metastasis. Nat Commun 2018;9:1793. [Crossref] [PubMed]
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426-37. [Crossref] [PubMed]
- Blakely CM, Watkins TBK, Wu W, et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet 2017;49:1693-704. [Crossref] [PubMed]
- Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov 2017;7:1394-403. [Crossref] [PubMed]
- Wu W, Huang Y, Guo J, et al. Detection and comparison of EGFR mutations from supernatants that contain cell-free DNA and cell pellets from FNA non-small cell lung cancer specimens. Cancer Cytopathol 2020;128:545-52. [Crossref] [PubMed]
- Southwood M, Krenz T, Cant N, et al. Systematic evaluation of PAXgene® tissue fixation for the histopathological and molecular study of lung cancer. J Pathol Clin Res 2020;6:40-54. [Crossref] [PubMed]
- Bellevicine C, Vita GD, Malapelle U, et al. Applications and limitations of oncogene mutation testing in clinical cytopathology. Semin Diagn Pathol 2013;30:284-97. [Crossref] [PubMed]
- Rolfo C, Mack PC, Scagliotti GV, et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J Thorac Oncol 2018;13:1248-68. [Crossref] [PubMed]
- Pisapia P, Malapelle U, Troncone G. Liquid Biopsy and Lung Cancer. Acta Cytol 2019;63:489-96. [Crossref] [PubMed]
- Siravegna G, Mussolin B, Venesio T, et al. How liquid biopsies can change clinical practice in oncology. Ann Oncol 2019;30:1580-90. [Crossref] [PubMed]
- Rijavec E, Coco S, Genova C, et al. Liquid Biopsy in Non-Small Cell Lung Cancer: Highlights and Challenges. Cancers (Basel) 2019.
- Revelo AE, Martin A, Velasquez R, et al. Liquid biopsy for lung cancers: an update on recent developments. Ann Transl Med 2019;7:349. [Crossref] [PubMed]
- Yi X, Ma J, Guan Y, et al. The feasibility of using mutation detection in ctDNA to assess tumor dynamics. Int J Cancer 2017;140:2642-7. [Crossref] [PubMed]
- Phallen J, Leal A, Woodward BD, et al. Early Noninvasive Detection of Response to Targeted Therapy in Non-Small Cell Lung Cancer. Cancer Res 2019;79:1204-13. [Crossref] [PubMed]
- Chae YK, Oh MS. Detection of Minimal Residual Disease Using ctDNA in Lung Cancer: Current Evidence and Future Directions. J Thorac Oncol 2019;14:16-24. [Crossref] [PubMed]
- Marrugo-Ramírez J, Mir M, et al. Blood-Based Cancer Biomarkers in Liquid Biopsy: A Promising Non-Invasive Alternative to Tissue Biopsy. Int J Mol Sci 2018;19:2877. [Crossref] [PubMed]
- Liu MC, Oxnard GR, Klein EA, et al. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol 2020;31:745-59. [Crossref] [PubMed]
- Fiala C, Diamandis EP. Utility of circulating tumor DNA in cancer diagnostics with emphasis on early detection. BMC Med 2018;16:166. [Crossref] [PubMed]
- Palmirotta R, Lovero D, Cafforio P, et al. Liquid biopsy of cancer: a multimodal diagnostic tool in clinical oncology. Ther Adv Med Oncol 2018;10:1758835918794630 [Crossref] [PubMed]
- Hoshino A, Kim HS, Bojmar L, et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell 2020;182:1044-61.e18. [Crossref] [PubMed]
- Oxnard GR, Thress KS, Alden RS, et al. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3375-82. [Crossref] [PubMed]
- Wu YL, Sequist LV, Hu CP, et al. EGFR mutation detection in circulating cell-free DNA of lung adenocarcinoma patients: analysis of LUX-Lung 3 and 6. Br J Cancer 2017;116:175-85. [Crossref] [PubMed]
- Tseng JS, Yang TY, Tsai CR, et al. Dynamic plasma EGFR mutation status as a predictor of EGFR-TKI efficacy in patients with EGFR-mutant lung adenocarcinoma. J Thorac Oncol 2015;10:603-10. [Crossref] [PubMed]
- Kasahara N, Kenmotsu H, Serizawa M, et al. Plasma epidermal growth factor receptor mutation testing with a chip-based digital PCR system in patients with advanced non-small cell lung cancer. Lung Cancer 2017;106:138-44. [Crossref] [PubMed]
- Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015;90:509-15. [Crossref] [PubMed]
- Jenkins S, Yang JC, Jänne PA, et al. EGFR Mutation Analysis for Prospective Patient Selection in Two Phase II Registration Studies of Osimertinib. J Thorac Oncol 2017;12:1247-56. [Crossref] [PubMed]
- Passiglia F, Rizzo S, Rolfo C, et al. Metastatic Site Location Influences the Diagnostic Accuracy of ctDNA EGFR- Mutation Testing in NSCLC Patients: a Pooled Analysis. Curr Cancer Drug Targets 2018;18:697-705. [Crossref] [PubMed]
- Villatoro S, Mayo-de-Las-Casas C, Jordana-Ariza N, et al. Prospective detection of mutations in cerebrospinal fluid, pleural effusion, and ascites of advanced cancer patients to guide treatment decisions. Mol Oncol 2019;13:2633-45. [Crossref] [PubMed]
- Iwama E, Sakai K, Hidaka N, et al. Longitudinal monitoring of somatic genetic alterations in circulating cell-free DNA during treatment with epidermal growth factor receptor-tyrosine kinase inhibitors. Cancer 2020;126:219-27. [Crossref] [PubMed]
- Lee JY, Qing X, Xiumin W, et al. Longitudinal monitoring of EGFR mutations in plasma predicts outcomes of NSCLC patients treated with EGFR TKIs: Korean Lung Cancer Consortium (KLCC-12-02). Oncotarget 2016;7:6984-93. [Crossref] [PubMed]
- Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res 2014;20:1698-705. [Crossref] [PubMed]
- Iwama E, Sakai K, Azuma K, et al. Monitoring of somatic mutations in circulating cell-free DNA by digital PCR and next-generation sequencing during afatinib treatment in patients with lung adenocarcinoma positive for EGFR activating mutations. Ann Oncol 2017;28:136-41. [Crossref] [PubMed]
- Malapelle U, Rolfo CInternational Society of Liquid Biopsy (ISLB). Liquid biopsy as a follow-up tool: Comment on longitudinal monitoring of somatic genetic alterations in circulating cell-free DNA during treatment with epidermal growth factor receptor-tyrosine kinase inhibitors. Cancer 2020;126:22-5. [Crossref] [PubMed]
- Zhou C, Imamura F, Cheng Y, et al. Early clearance of plasma EGFR mutations as a predictor of response to osimertinib and comparator EGFR-TKIs in the FLAURA trial. J Clin Oncol 2019;37:9020. [Crossref]
- Thress KS, Markovets A, Barrett JC, et al. Complete clearance of plasma EGFR mutations as a predictor of outcome on osimertinib in the AURA trial. J Clin Oncol 2017;35:9018. [Crossref]
- Shepherd FA, Papadimitrakopoulou V, Mok T, et al. Early clearance of plasma EGFR mutations as a predictor of response to osimertinib in the AURA3 trial. J Clin Oncol 2018;36:9027. [Crossref]
- Anagnostou V, Forde PM, White JR, et al. Dynamics of Tumor and Immune Responses during Immune Checkpoint Blockade in Non-Small Cell Lung Cancer. Cancer Res 2019;79:1214-25. [Crossref] [PubMed]
- Bratman SV, Yang SYC, Iafolla MAJ, et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nature Cancer 2020;1:873-81. [Crossref]
- Goldberg SB, Narayan A, Kole AJ, et al. Early Assessment of Lung Cancer Immunotherapy Response via Circulating Tumor DNA. Clin Cancer Res 2018;24:1872-80. [Crossref] [PubMed]
- Zhang Q, Luo J, Wu S, et al. Prognostic and Predictive Impact of Circulating Tumor DNA in Patients with Advanced Cancers Treated with Immune Checkpoint Blockade. Cancer Discov 2020;10:1842-53. [Crossref] [PubMed]
- Tang C, Lee WC, Reuben A, et al. Immune and Circulating Tumor DNA Profiling After Radiation Treatment for Oligometastatic Non-Small Cell Lung Cancer: Translational Correlatives from a Mature Randomized Phase II Trial. Int J Radiat Oncol Biol Phys 2020;106:349-57. [Crossref] [PubMed]
- Chabon JJ, Hamilton EG, Kurtz DM, et al. Integrating genomic features for non-invasive early lung cancer detection. Nature 2020;580:245-51. [Crossref] [PubMed]
- Rolfo C, Russo A. Liquid biopsy for early stage lung cancer moves ever closer. Nat Rev Clin Oncol 2020;17:523-4. [Crossref] [PubMed]
Systemic Therapy With or Without Local Consolidative Therapy in Treating Patients With Oligometastatic Solid Tumor Nivolumab and Ipilimumab With or Without Local Consolidation Therapy in Treating Patients With Stage IV Non-Small Cell Lung Cancer - Banna GL, Passiglia F, Colonese F, et al. Immune-checkpoint inhibitors in non-small cell lung cancer: A tool to improve patients' selection. Crit Rev Oncol Hematol 2018;129:27-39. [Crossref] [PubMed]
- Addeo A, Banna GL, Weiss GJ. Tumor Mutation Burden-From Hopes to Doubts. JAMA Oncol 2019;5:934-5. [Crossref] [PubMed]
- Munari E, Zamboni G, Lunardi G, et al. PD-L1 Expression Heterogeneity in Non-Small Cell Lung Cancer: Defining Criteria for Harmonization between Biopsy Specimens and Whole Sections. J Thorac Oncol 2018;13:1113-20. [Crossref] [PubMed]
- Mildner F, Sopper S, Amann A, et al. Systematic review: Soluble immunological biomarkers in advanced non-small-cell lung cancer (NSCLC). Crit Rev Oncol Hematol 2020;153:102948 [Crossref] [PubMed]
- Wang Z, Duan J, Cai S, et al. Assessment of Blood Tumor Mutational Burden as a Potential Biomarker for Immunotherapy in Patients With Non-Small Cell Lung Cancer With Use of a Next-Generation Sequencing Cancer Gene Panel. JAMA Oncol 2019;5:696-702. [Crossref] [PubMed]
- Gandara DR, Paul SM, Kowanetz M, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med 2018;24:1441-8. [Crossref] [PubMed]
- Iijima Y, Hirotsu Y, Amemiya K, et al. Very early response of circulating tumour-derived DNA in plasma predicts efficacy of nivolumab treatment in patients with non-small cell lung cancer. Eur J Cancer 2017;86:349-57. [Crossref] [PubMed]
- Raja R, Kuziora M, Brohawn PZ, et al. Early Reduction in ctDNA Predicts Survival in Patients with Lung and Bladder Cancer Treated with Durvalumab. Clin Cancer Res 2018;24:6212-22. [Crossref] [PubMed]
- Frick MA, Feigenberg SJ, Jean-Baptiste SR, et al. Circulating Tumor Cells Are Associated with Recurrent Disease in Patients with Early-Stage Non-Small Cell Lung Cancer Treated with Stereotactic Body Radiotherapy. Clin Cancer Res 2020;26:2372-80. [Crossref] [PubMed]
- Chinniah C, Aguarin L, Cheng P, et al. Early Detection of Recurrence in Patients With Locally Advanced Non-Small-Cell Lung Cancer via Circulating Tumor Cell Analysis. Clin Lung Cancer 2019;20:384-390.e2. [Crossref] [PubMed]
- Hanssen A, Riebensahm C, Mohme M, et al. Frequency of Circulating Tumor Cells (CTC) in Patients with Brain Metastases: Implications as a Risk Assessment Marker in Oligo-Metastatic Disease. Cancers (Basel) 2018.
- Sullivan R, Maresh G, Zhang X, et al. The Emerging Roles of Extracellular Vesicles As Communication Vehicles within the Tumor Microenvironment and Beyond. Front Endocrinol (Lausanne) 2017;8:194. [Crossref] [PubMed]
- Chen G, Huang AC, Zhang W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 2018;560:382-6. [Crossref] [PubMed]
- Del Re M, Marconcini R, Pasquini G, et al. PD-L1 mRNA expression in plasma-derived exosomes is associated with response to anti-PD-1 antibodies in melanoma and NSCLC. Br J Cancer 2018;118:820-4. [Crossref] [PubMed]
- Okuma Y, Wakui H, Utsumi H, et al. Soluble Programmed Cell Death Ligand 1 as a Novel Biomarker for Nivolumab Therapy for Non-Small-cell Lung Cancer. Clin Lung Cancer 2018;19:410-417.e1. [Crossref] [PubMed]
- Costantini A, Julie C, Dumenil C, et al. Predictive role of plasmatic biomarkers in advanced non-small cell lung cancer treated by nivolumab. Oncoimmunology 2018;7:e1452581 [Crossref] [PubMed]
- Chen Y, Wang Q, Shi B, et al. Development of a sandwich ELISA for evaluating soluble PD-L1 (CD274) in human sera of different ages as well as supernatants of PD-L1+ cell lines. Cytokine 2011;56:231-8. [Crossref] [PubMed]
- Zhao J, Zhang P, Wang J, et al. Plasma levels of soluble programmed death ligand-1 may be associated with overall survival in nonsmall cell lung cancer patients receiving thoracic radiotherapy. Medicine (Baltimore) 2017;96:e6102 [Crossref] [PubMed]
- Sorensen SF, Demuth C, Weber B, et al. Increase in soluble PD-1 is associated with prolonged survival in patients with advanced EGFR-mutated non-small cell lung cancer treated with erlotinib. Lung Cancer 2016;100:77-84. [Crossref] [PubMed]
- Corkum CP, Ings DP, Burgess C, et al. Immune cell subsets and their gene expression profiles from human PBMC isolated by Vacutainer Cell Preparation Tube (CPT™) and standard density gradient. BMC Immunol 2015;16:48. [Crossref] [PubMed]
- Zheng H, Liu X, Zhang J, et al. Expression of PD-1 on CD4+ T cells in peripheral blood associates with poor clinical outcome in non-small cell lung cancer. Oncotarget 2016;7:56233-40. [Crossref] [PubMed]
- Kamphorst AO, Pillai RN, Yang S, et al. Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1-targeted therapy in lung cancer patients. Proc Natl Acad Sci U S A 2017;114:4993-8. [Crossref] [PubMed]
- Mazzaschi G, Facchinetti F, Missale G, et al. The circulating pool of functionally competent NK and CD8+ cells predicts the outcome of anti-PD1 treatment in advanced NSCLC. Lung Cancer 2019;127:153-63. [Crossref] [PubMed]
- Tavakkoli M, Wilkins CR, Mones JV, et al. A Novel Paradigm Between Leukocytosis, G-CSF Secretion, Neutrophil-to-Lymphocyte Ratio, Myeloid-Derived Suppressor Cells, and Prognosis in Non-small Cell Lung Cancer. Front Oncol 2019;9:295. [Crossref] [PubMed]
- Bagley SJ, Kothari S, Aggarwal C, et al. Pretreatment neutrophil-to-lymphocyte ratio as a marker of outcomes in nivolumab-treated patients with advanced non-small-cell lung cancer. Lung Cancer 2017;106:1-7. [Crossref] [PubMed]
- Mezquita L, Auclin E, Ferrara R, et al. Association of the Lung Immune Prognostic Index With Immune Checkpoint Inhibitor Outcomes in Patients With Advanced Non-Small Cell Lung Cancer. JAMA Oncol 2018;4:351-7. [Crossref] [PubMed]
- Banna GL, Signorelli D, Metro G, et al. Neutrophil-to-lymphocyte ratio in combination with PD-L1 or lactate dehydrogenase as biomarkers for high PD-L1 non-small cell lung cancer treated with first-line pembrolizumab. Transl Lung Cancer Res 2020;9:1533-42. [Crossref] [PubMed]
- Remon J, Menis J, Hasan B, et al. The APPLE Trial: Feasibility and Activity of AZD9291 (Osimertinib) Treatment on Positive PLasma T790M in EGFR-mutant NSCLC Patients. EORTC 1613. Clin Lung Cancer 2017;18:583-8. [Crossref] [PubMed]
- Banna GL, Olivier T, Rundo F, et al. The Promise of Digital Biopsy for the Prediction of Tumor Molecular Features and Clinical Outcomes Associated With Immunotherapy. Front Med (Lausanne) 2019;6:172. [Crossref] [PubMed]
- Chin AL, Kumar KA, Guo HH, et al. Prognostic Value of Pretreatment FDG-PET Parameters in High-dose Image-guided Radiotherapy for Oligometastatic Non-Small-cell Lung Cancer. Clin Lung Cancer 2018;19:e581-8. [Crossref] [PubMed]
- Papadimitrakopoulou VA, Mok TS, Han JY, et al. Osimertinib versus platinum-pemetrexed for patients with EGFR T790M advanced NSCLC and progression on a prior EGFR-tyrosine kinase inhibitor: AURA3 overall survival analysis. Ann Oncol 2020;31:1536-44. [Crossref] [PubMed]
- Ettinger DS, Wood DE, Aisner DL, et al. Non-Small Cell Lung Cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2017;15:504-35. [Crossref] [PubMed]
- Papadimitrakopoulou VA, Han JY, Ahn MJ, et al. Epidermal growth factor receptor mutation analysis in tissue and plasma from the AURA3 trial: Osimertinib versus platinum-pemetrexed for T790M mutation-positive advanced non-small cell lung cancer. Cancer 2020;126:373-80. [Crossref] [PubMed]
- Zhao J, Ye X, Xu Y, et al. EGFR mutation status of paired cerebrospinal fluid and plasma samples in EGFR mutant non-small cell lung cancer with leptomeningeal metastases. Cancer Chemother Pharmacol 2016;78:1305-10. [Crossref] [PubMed]
- Xing L, Pan Y, Shi Y, et al. Biomarkers of Osimertinib Response in Patients with Refractory, EGFR-T790M-positive Non-Small Cell Lung Cancer and Central Nervous System Metastases: The APOLLO Study. Clin Cancer Res 2020;26:6168-75. [Crossref] [PubMed]
- Hu M, Wu YL, Zhu X, et al. Identification of osimertinib resistance mechanisms in Chinese NSCLC patients: Analysis from AURA17 trial. J Clin Oncol 2018;35:9077. [Crossref]
- Ni J, Weng L, Liu Y, et al. Dynamic monitoring of EGFR mutations in circulating cell-free DNA for EGFR-mutant metastatic patients with lung cancer: Early detection of drug resistance and prognostic significance. Oncol Lett 2017;13:4549-57. [Crossref] [PubMed]
- Zhang EW, Dagogo-Jack I, Kuo A, et al. Association between circulating tumor DNA burden and disease burden in patients with ALK-positive lung cancer. Cancer 2020;126:4473-84. [Crossref] [PubMed]
- Aldea M, Hendriks L, Mezquita L, et al. Circulating Tumor DNA Analysis for Patients with Oncogene-Addicted NSCLC With Isolated Central Nervous System Progression. J Thorac Oncol 2020;15:383-91. [Crossref] [PubMed]
- Zheng MM, Li YS, Jiang BY, et al. Clinical Utility of Cerebrospinal Fluid Cell-Free DNA as Liquid Biopsy for Leptomeningeal Metastases in ALK-Rearranged NSCLC. J Thorac Oncol 2019;14:924-32. [Crossref] [PubMed]
- Madison R, Schrock AB, Castellanos E, et al. Retrospective analysis of real-world data to determine clinical outcomes of patients with advanced non-small cell lung cancer following cell-free circulating tumor DNA genomic profiling. Lung Cancer 2020;148:69-78. [Crossref] [PubMed]
- Moding EJ, Liu Y, Nabet BY, et al. Circulating tumor DNA dynamics predict benefit from consolidation immunotherapy in locally advanced non-small-cell lung cancer. Nat Cancer 2020;1:176-83. [Crossref]
- Aggarwal C, Rolfo CD, Oxnard GR, et al. Strategies for the successful implementation of plasma-based NSCLC genotyping in clinical practice. Nat Rev Clin Oncol 2021;18:56-62. [Crossref] [PubMed]


