
Blood digital polymerase chain reaction as a potential method to detect human epidermal growth factor receptor 2 amplification in non-small cell lung cancer
Introduction
Lung cancer is one of the most lethal cancers and is a serious threat to human life (1). Non-small cell lung cancer (NSCLC) accounts for 80% of all lung cancers (2), and the 5-year survival rate among all NSCLC patients is about 10% (3). Thus, it is necessary to improve the clinical diagnosis and prognosis of NSCLC.
Human epidermal growth factor receptor 2 (HER2) is a member of the epidermal growth factor receptor (EGFR) subfamily (4) and HER2 amplification has been detected in NSCLC (5). Previous studies have shown that HER2 gene amplification was present in 10–20% of NSCLC patients (6,7). Notably, HER2 amplification rates have been shown to be associated with the drug resistance in NSCLC (8,9). At present, the clinical detection of HER2 amplification is generally by immunohistochemistry (IHC) or fluorescence in situ hybridization (FISH) (10). However, IHC and FISH have some limitations, including difficulty in obtaining adequate samples (11).
Digital polymerase chain reaction (dPCR), a new nucleic acid detection technology, can achieve quantitative analysis of target nucleic acid molecules and accurately analyze the target gene copy number variation in tumor tissue or blood at the nucleic acid level (12). It could make sample dispersed and diluted resulting to statistically one or no DNA molecule in each chamber for further amplification instead of performing amplification in bulk sample as traditional polymerase chain reaction (PCR) (13). Currently, the sample dispersion method could be droplet-based, microwell-base, channel-based, hydrogel-based and printing-based (13). This technique could avoid the signal of rare genetic changes being ignored and largely improving the sensitivity and precision of detecting rare genetic aberrations. Previous studies have demonstrated that the accuracy of this technique to detected EGFR and KRAS mutation through circulating tumor DNA (ctDNA) in NSCLC (14,15). However, whether this technique could detect the amplification of HER2 in NSCLC by plasma genotyping had not been discussed previously. Besides dPCR, next-generation sequencing (NGS) is also used to reveal HER2 alterations in NSCLC patients (16). However, compared with NGS, the validity of dPCR, and the feasibility of detecting in blood samples in NSCLC patients are still unclear.
In this study, a comparative analysis of dPCR and NGS was performed regarding their efficacy in HER2 amplification detection in fresh tissues and blood samples of NSCLC patients. We verified the feasibility of dPCR detection compared with NGS and explored the possibility of detecting NSCLC HER2 amplification in blood samples instead of tissue samples. We present the following article in accordance with the STARD reporting checklist (available at https://dx.doi.org/10.21037/tlcr-21-860).
Methods
Patients
As we would like to evaluate the efficacy of dPCR in detecting HER2 amplification through tissue and blood samples, we involved a total of 112 biopsy tissues and 88 blood samples from NSCLC patients who were treated from November 2017 to May 2019 in Shanghai Pulmonary hospital to have relevant test. Clinical information was collected from each participant. Histological diagnoses were made according to the WHO classification (17), and the stages were classed according to the International Association for the Study of Lung Cancer (IASLC) 8th edition (18). Patients who were diagnosed as NSCLC and older than 18 years old could be involved. Written consent was provided by all participants. The study was approved by the Ethics Committee of Shanghai Pulmonary Hospital (No. K20-275). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The patients’ characteristics would be blinded for researchers who performed the test.
Extraction of serum ctDNA
The ctDNA of blood samples was extracted using a QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany).
DNA extraction from tissue sample
DNA was extracted from fresh tissue samples preserved in RNAlater. DNA extraction was performed with the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacture’s recommendation.
HER2 gene copy number evaluation by dPCR
The copy number variation of the HER2 gene in the genomic DNA (gDNA) of all tissues and ctDNA of blood samples was detected by dPCR. Based on Beacon DesignerTM 8.12 software (Premier Biosoft, Palo Alto, CA, USA) and the human HER2 gene sequence, the sequence-specific oligonucleotide primer and TaqMan probe were designed to detect HER2 and elongation factor Tu GTP binding domain containing 2 (EFTUD2) of the internal reference gene. All the primers were synthesized by Shanghai Biotechnology Co., Ltd. (Shanghai, China). The cycling conditions were the same as previously described (19). The sequence of primers were listed below: HER2-F: 5'-CTGCGGATTGTGCGAGG-3'; HER2-R: 5'-CAGCGGGTCTCCATTGTC-3'; HER2-probe: 5'-CCCAGCTCTTTGAGGACAAC-3'; EFTUD2-F: 5'-CTCTTCAATATCATGGACACTCCAG-3'; EFTUD2-R: 5'-CGCAAAACCAAGACAAGGTTC-3'; EFTUD2-probe: 5'-GGACATCCTTTGGCTTTTGA-3' (Table S1).
HER2 gene copy number evaluation by NGS
The copy number variation of the HER2 gene in the gDNA of all tissue samples and ctDNA of blood samples was detected by NGS. The gDNA with a double terminal 8-base UDI connector and ctDNA with a single terminal 8-base plus 8-base UMI internal connector was used for the library construction, respectively. Then, the library was hybridized with an Agilent SureSelectQXT reagent kit and blocking agent (Agilent Technologies, Santa Clara, CA, USA). Finally, PCR was used to enrich the captured target bands, and the library was prepared after the capture. After fragment size quantitation by Qubit 4.0 (Thermo Fisher, Waltham, MA, USA) concentration and 4200 Bioanalyzer (Agilent), the library was quantified by quantitative polymerase chain reaction (qPCR), mixed, and then sequenced on NovaSeq 6000 platform (Illumina, San Diego, CA, USA; 4 g tissue data, 15 g blood data). The original data obtained after sequencing was automatically converted into FASTQ data using bcl2fastq software (Illumina) for subsequent data analysis.
The optimal cutoff value for dPCR
The cutoff value of dPCR was determined according to the NGS result. For the tissue samples, when the amplification frequency of HER2 detected by dPCR was ≥61.38% (amplification multiple ≥1.59 times), it was consistent with that detected by NGS (amplification multiple ≥1.42). For blood (plasma) samples, when the amplification frequency of HER2 detected by dPCR was ≥58.81% (amplification multiple ≥1.43 times), it was consistent with that detected by NGS (amplification multiple ≥1.45 times).
Statistical analyses
Data were analyzed using the software SAS 9.0 (SAS Institute, Cary, NC, USA). Indeterminate result of test would not be included for further analysis. All data were represented as the mean ± standard deviation (SD). Student’s t-test was used to analyze continuous variables, and chi-square tests were used to analyze categorical variables. Moreover, the survival analysis was performed based on the Kaplan-Meier (KM) method. The log-rank test was used to calculate statistical differences in survival status.
Furthermore, the sensitivity and specificity analysis of each method was performed based on the ROC curve. The Cox proportional hazards model was used in the regression analysis. A P value <0.05 was considered statistically significant.
Results
Participant characteristics
We recruited 112 participants (59 male; 53 female) to the tissue sample group and detected HER2 with dPCR. Among them, 36 were smokers and 76 were non-smokers. A total of 63 participants had EGFR mutations, and 51 had received at least first-line therapy with tyrosine kinase inhibitors (TKIs) (Table 1).
Table 1
| Factors | Negative group (n=63), n (%) | Positive group (n=49), n (%) | χ2 | P value |
|---|---|---|---|---|
| Sex | 0.205 | 0.651 | ||
| Male | 32 (50.8) | 27 (55.1) | ||
| Female | 31 (49.2) | 22 (44.9) | ||
| Age (years) | 0.291 | 0.590 | ||
| <60 | 21 (33.3) | 14 (28.6) | ||
| ≥60 | 42 (66.7) | 35 (71.4) | ||
| Smoking | 1.757 | 0.185 | ||
| No | 46 (73.0) | 30 (61.2) | ||
| Yes | 17 (27.0) | 19 (38.8) | ||
| NSCLC types | 9.253 | 0.010 | ||
| ac-NSCLC | 45 (71.4) | 28 (57.1) | ||
| sq-NSCLC | 3 (4.8) | 12 (24.5) | ||
| NSCLC | 15 (23.8) | 9 (18.4) | ||
| TNM stage | 0.054 | 0.816 | ||
| III | 13 (20.6) | 11 (22.4) | ||
| IV | 50 (76.4) | 38 (77.6) | ||
| Bone metastasis | 0.260 | 0.610 | ||
| No | 44 (69.8) | 32 (65.3) | ||
| Yes | 19 (30.2) | 17 (34.7) | ||
| Pulmonary metastasis | 1.418 | 0.234 | ||
| No | 41 (65.1) | 37 (75.5) | ||
| Yes | 22 (34.9) | 12 (24.5) | ||
| Brain metastases | 0.139 | 0.709 | ||
| No | 51 (81.0) | 41 (83.7) | ||
| Yes | 12 (19.0) | 8 (16.3) | ||
| Pleura metastases | 0.361 | 0.548 | ||
| No | 43 (68.3) | 36 (73.5) | ||
| Yes | 20 (31.7) | 13 (26.5) | ||
| Adrenal gland metastases | 2.885 | 0.089 | ||
| No | 62 (98.4) | 45 (91.8) | ||
| Yes | 1 (1.6) | 4 (8.2) | ||
| Liver metastases | 0.138 | 0.710 | ||
| No | 58 (92.1) | 46 (93.9) | ||
| Yes | 5 (7.9) | 3 (6.1) | ||
| LN metastases | 0.065 | 0.798 | ||
| No | 61 (96.8) | 47 (95.9) | ||
| Yes | 2 (3.2) | 2 (4.1) | ||
| EGFR mutation | 0.888 | 0.642 | ||
| Common | 34 (79.1) | 16 (69.6) | ||
| Uncommon | 7 (16.3) | 6 (26.1) | ||
| WT | 2 (4.7) | 1 (4.3) | ||
| TKI | 2.776 | 0.096 | ||
| No | 21 (38.9) | 23 (56.1) | ||
| Yes | 33 (61.1) | 18 (43.9) |
P<0.05 was considered as significant different. NSCLC, non-small cell lung cancer; HER2, human epidermal growth factor receptor-2; dPCR, digital polymerase chain reaction; ac-NSCLC, lung adenocarcinoma; sq-NSCLC, lung squamous cell carcinoma; TNM, tumor, lymph node and metastasis; LN, Lymph node; EGFR, epidermal growth factor receptor; WT, wide type; TKI, Tyrosine kinase inhibitors.
A total of 90 participants in the tissue sample group were also detected by NGS (46 male; 44 female). A total of 29 were smokers, 61 were non-smokers; 12 were squamous and 78 non-squamous; 52 participants had EGFR mutations, and 42 had received at least first-line therapy with TKIs (Table 2).
Table 2
| Factors | Negative group (n=44), n (%) | Positive group (n=46), n (%) | χ2 | P value |
|---|---|---|---|---|
| Sex | 2.166 | 0.141 | ||
| Male | 19 (43.2) | 27 (58.7) | ||
| Female | 25 (56.8) | 19 (41.3) | ||
| Age (years) | 2.233 | 0.136 | ||
| <60 | 18 (40.9) | 12 (26.1) | ||
| ≥60 | 26 (59.1) | 34 (73.9) | ||
| Smoking | 2.056 | 0.152 | ||
| No | 33 (75.0) | 28 (60.9) | ||
| Yes | 11 (25.0) | 18 (39.1) | ||
| NSCLC types | 5.570 | 0.062 | ||
| ac-NSCLC | 33 (75.0) | 24 (52.2) | ||
| sq-NSCLC | 3 (6.8) | 9 (19.6) | ||
| NSCLC | 8 (18.2) | 13 (28.3) | ||
| TNM stage | 1.985 | 0.159 | ||
| III | 7 (15.9) | 13 (28.3) | ||
| IV | 37 (84.1) | 33 (71.7) | ||
| Bone metastasis | 5.852 | 0.016 | ||
| No | 25 (56.8) | 37 (80.4) | ||
| Yes | 19 (43.2) | 9 (19.6) | ||
| Pulmonary metastasis | 0.357 | 0.550 | ||
| No | 29 (65.9) | 33 (71.7) | ||
| Yes | 15 (34.1) | 13 (28.3) | ||
| Brain metastases | 0.028 | 0.867 | ||
| No | 36 (81.8) | 37 (80.4) | ||
| Yes | 8 (18.2) | 9 (19.6) | ||
| Pleura metastases | 1.660 | 0.198 | ||
| No | 28 (63.6) | 35 (76.1) | ||
| Yes | 16 (36.4) | 11 (23.9) | ||
| Adrenal gland metastases | 0.399 | 0.528 | ||
| No | 42 (95.5) | 45 (97.8) | ||
| Yes | 2 (4.5) | 1 (2.2) | ||
| Liver metastases | 3.296 | 0.069 | ||
| No | 39 (88.6) | 45 (97.8) | ||
| Yes | 5 (11.4) | 1 (2.2) | ||
| LN metastases | 1.188 | 0.276 | ||
| No | 41 (93.2) | 45 (97.8) | ||
| Yes | 3 (6.8) | 1 (2.2) | ||
| EGFR mutation | 5.597 | 0.061 | ||
| Common | 26 (81.3) | 17 (73.9) | ||
| Uncommon | 3 (9.4) | 6 (26.1) | ||
| WT | 3 (9.4) | 0 (0.0) | ||
| TKI | 0.315 | 0.574 | ||
| No | 20 (47.6) | 14 (41.2) | ||
| Yes | 22 (52.4) | 20 (58.8) |
P<0.05 was considered as significant different. NSCLC, non-small cell lung cancer; HER2, human epidermal growth factor receptor-2; dPCR, digital polymerase chain reaction; ac-NSCLC, lung adenocarcinoma; sq-NSCLC, lung squamous cell carcinoma; TNM, tumor, lymph node and metastasis; LN, Lymph node; EGFR, epidermal growth factor receptor; WT, wide type; TKI, Tyrosine kinase inhibitors.
A total of 88 participants (46 male; 42 female) were enrolled and had HER2 detected by dPCR in their blood samples. Among them, 32 were smokers and 56 were non-smokers (Table 3). Sixteen blood samples were also analyzed by NGS and patients’ characteristics were summarized in Table 4.
Table 3
| Factors | Negative group (n=42), n (%) | Positive group (n=46), n (%) | χ2 | P value |
|---|---|---|---|---|
| Sex | 0.697 | 0.404 | ||
| Male | 20 (47.6) | 26 (56.5) | ||
| Female | 22 (52.4) | 20 (43.5) | ||
| Age (years) | 0.573 | 0.449 | ||
| <60 | 16 (38.1) | 14 (30.4) | ||
| ≥60 | 26 (61.9) | 32 (69.6) | ||
| Smoking | 0.015 | 0.904 | ||
| No | 27 (64.3) | 29 (63.0) | ||
| Yes | 15 (35.7) | 17 (37.0) | ||
| NSCLC types | 0.441 | 0.802 | ||
| ac-NSCLC | 27 (64.3) | 29 (63.0) | ||
| sq-NSCLC | 7 (16.7) | 6 (13.0) | ||
| NSCLC | 8 (19.0) | 11 (23.9) | ||
| TNM stage | 1.004 | 0.316 | ||
| III | 11 (26.2) | 8 (17.4) | ||
| IV | 31 (73.8) | 38 (82.6) | ||
| Bone metastasis | 0.554 | 0.457 | ||
| No | 28 (66.7) | 34 (73.9) | ||
| Yes | 14 (33.3) | 12 (26.1) | ||
| Pulmonary metastasis | 0.003 | 0.958 | ||
| No | 29 (69.0) | 32 (69.6) | ||
| Yes | 13 (31.0) | 14 (30.4) | ||
| Brain metastases | 0.709 | 0.400 | ||
| No | 35 (83.3) | 35 (86.1) | ||
| Yes | 7 (16.7) | 11 (23.9) | ||
| Pleura metastases | 0.047 | 0.828 | ||
| No | 31 (73.8) | 33 (71.7) | ||
| Yes | 11 (26.2) | 13 (28.3) | ||
| Adrenal gland metastases | 0.128 | 0.721 | ||
| No | 40 (95.2) | 43 (93.5) | ||
| Yes | 2 (4.8) | 3 (6.5) | ||
| Liver metastases | 1.758 | 0.185 | ||
| No | 41 (97.6) | 42 (91.3) | ||
| Yes | 1 (2.4) | 4 (8.7) | ||
| LN metastases | 0.264 | 0.608 | ||
| No | 41 (97.6) | 44 (95.7) | ||
| Yes | 1 (2.4) | 2 (4.3) | ||
| EGFR mutation | 2.557 | 0.278 | ||
| Common | 17 (70.8) | 21 (80.8) | ||
| Uncommon | 7 (29.2) | 4 (15.4) | ||
| WT | 0 (0.0) | 1 (3.8) | ||
| TKI | 0.001 | 0.979 | ||
| No | 16 (47.1) | 18 (47.4) | ||
| Yes | 18 (52.9) | 20 (52.6) |
P<0.05 was considered as significant different. NSCLC, non-small cell lung cancer; HER2, human epidermal growth factor receptor-2; dPCR, digital polymerase chain reaction; ac-NSCLC, lung adenocarcinoma; sq-NSCLC, lung squamous cell carcinoma; TNM, tumor, lymph node and metastasis; LN, lymph node; EGFR, epidermal growth factor receptor; WT, wide type; TKI, tyrosine kinase inhibitors.
Table 4
| Factors | Negative group (n=13), n (%) | Positive group (n=3), n (%) | χ2 | P value |
|---|---|---|---|---|
| Sex | 0.028 | 0.868 | ||
| Male | 5 (38.5) | 1 (33.3) | ||
| Female | 8 (61.5) | 2 (66.7) | ||
| Age (years) | 0.417 | 0.519 | ||
| <60 | 7 (53.8) | 1 (33.3) | ||
| ≥60 | 6 (46.2) | 2 (66.7) | ||
| Smoking | 0.461 | 0.497 | ||
| No | 11 (84.6) | 2 (66.7) | ||
| Yes | 2 (15.4) | 1 (33.3) | ||
| NSCLC types | 0.894 | 0.344 | ||
| ac-NSCLC | 11 (84.6) | 3 (100.0) | ||
| NSCLC | 2 (15.4) | 0 (0.0) | ||
| TNM stage | 0.430 | 0.512 | ||
| III | 1 (7.7) | 0 (0.0) | ||
| IV | 12 (92.3) | 3 (100.0) | ||
| Bone metastasis | 3.225 | 0.073 | ||
| No | 7 (53.8) | 3 (100.0) | ||
| Yes | 6 (46.2) | 0 (0.0) | ||
| Pulmonary metastasis | 3.013 | 0.083 | ||
| No | 11 (84.6) | 1 (33.3) | ||
| Yes | 2 (15.4) | 2 (66.7) | ||
| Brain metastases | 0.130 | 0.718 | ||
| No | 10 (76.9) | 2 (66.7) | ||
| Yes | 3 (23.1) | 1 (33.3) | ||
| Pleura metastases | 3.225 | 0.073 | ||
| No | 7 (53.8) | 3 (100.0) | ||
| Yes | 6 (46.2) | 0 (0.0) | ||
| Adrenal gland metastases | – | – | ||
| No | 13 (100.0) | 3 (100.0) | ||
| Yes | 0 (0.0) | 0 (0.0) | ||
| Liver metastases | 0.461 | 0.497 | ||
| No | 11 (84.6) | 2 (66.7) | ||
| Yes | 2 (15.4) | 1 (33.3) | ||
| LN metastases | 3.662 | 0.056 | ||
| No | 13 (100.0) | 2 (66.7) | ||
| Yes | 0 (0.0) | 1 (33.3) | ||
| EGFR mutation | – | – | ||
| Common | 10 (100.0) | 1 (100.0) | ||
| Uncommon | 0 (0.0) | 0 (0.0) | ||
| WT | 0 (0.0) | 0 (0.0) | ||
| TKI | 0.200 | 0.655 | ||
| No | 4 (33.3) | 1 (50.0) | ||
| Yes | 8 (66.7) | 1 (50.0) |
P<0.05 was considered as significant different. NSCLC, non-small cell lung cancer; HER2, human epidermal growth factor receptor-2; NGS, next generation sequencing; ac-NSCLC, lung adenocarcinoma; TNM, tumor, lymph node and metastasis; LN, lymph node; EGFR, epidermal growth factor receptor; WT, wide type; TKI, tyrosine kinase inhibitors.
The sensitivity and specificity analysis based on ROC
The ROC analysis for the tissue sample (or blood samples) detected by dPCR (or NGS) is shown in Figure 1.
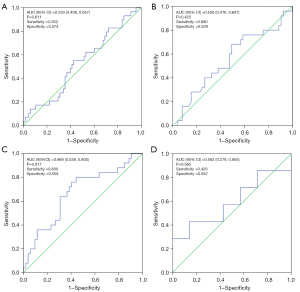
The results showed that the area under the curve (AUC) for tissue dPCR was 0.533 [95% confidence interval (CI): 0.408 to 0.657; P=0.611] with a sensitivity of 55.2% and a specificity of 57.4% (Figure 1A). The AUC for tissue NGS was 0.556 (95% CI: 0.416 to 0.697; P=0.425) with a sensitivity of 68.0% and a specificity of 50.9% (Figure 1B).
The results showed that the AUC for blood dPCR was 0.669 (95% CI: 0.538 to 0.800; P=0.017) with a sensitivity of 80.0% and a specificity of 55.8% (Figure 1C). Meanwhile, the AUC for blood NGS was 0.592 (95% CI: 0.279 to 0.905; P=0.565) with a sensitivity of 42.9% and a specificity of 85.7% (Figure 1D). The P values in blood dPCR were less than 0.05, indicating a good prediction result.
HER2 amplification in tissue samples and blood samples
When HER2 amplification of tissue samples was ≥1.59 times and HER2 amplification of blood samples ≥1.42 times, the results of paired detection of tissue and blood were consistent: 2/3 =66.7%. When the amplification of HER2 in tissue samples was <1.59 times and that of HER2 in blood samples <1.42 times, the results of dPCR match detection on tissue and blood samples were consistent: 96/97 =98.9%.
Correlation between HER2 amplification results and clinical data
Based on the tissue samples, we analyzed the correlation between clinical factors and HER2 amplification detected by dPCR and NGS in NSCLC patients and dPCR analysis showed that pathological type was correlated with HER2 amplification (P=0.010, Table 1). NGS analysis showed that bone metastasis was associated with HER2 amplification (P=0.016, Table 2).
Survival analysis
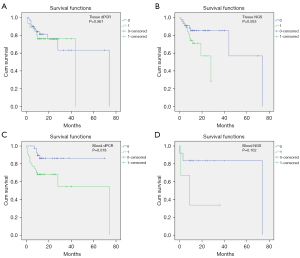
The results of univariate Cox regression analysis indicated that baseline information such as age [≥60 vs. <60, hazard ratio (HR) (95% CI): 4.621 (1.379 to 15.488), P=0.013], pleura metastasis [yes vs. no, HR (95% CI): 0.274 (0.082 to 0.920), P=0.036], and NSCLC type [adenocarcinoma vs. squamous cell carcinoma, HR (95% CI): 2.153 (1.361 to 3.405), P=0.001] were significantly associated with OS (Table S2). In addition, KM survival analysis showed that the differences between OS and HER2 amplification detected by tissue NGS (positive vs. negative, HR (95% CI): 55.38 (39.26 to 71.50), P=0.053) and blood dPCR (positive vs. negative, HR (95% CI): 20.61 (16.95 to 24.26), P=0.044).
In tissue dPCR, median OS of the negative group and positive group was 39.00 and 66.01 months, respectively (P=0.561) (Figure 2A). In tissue NGS, median survival time of the negative group and the positive group was 39.26 and 71.50 months, respectively, with a marginal difference (P=0.053) (Figure 2B). Moreover, in blood dPCR, the median survival time of the negative HER2 amplification group and the positive group was 61.67 (54.84 to 68.49) months and 45.44 (31.16 to 59.71) months, respectively (P=0.018) (Figure 2C). Multivariate Cox regression analysis showed that the risk of death in the blood dPCR HER2 amplification positive group was significantly higher than that in the negative group [HR (95% CI): 3.874 (1.356 to 11.069), P=0.011] (Table S3). Moreover, the multivariate Cox regression analysis of tissue NGS showed that after adjusting for factors such as age, pleura metastasis, NSCLC pathology type, and smoking, HER2 amplification was not significantly associated with the risk of death (Table S4). Furthermore, in blood NGS, the median survival time of the negative HER2 amplification group and the positive group was 61.92 (43.19 to 80.65) months and 15.33 (0 to 32.28), respectively (P=0.102) (Figure 2D).
Discussion
As the most common type of lung cancer, the incidence of NSCLC is high (20). Although dPCR is an approach for detecting biomarkers in cancer, whether dPCR is feasible for HER2 amplification is unknown. In this study, we used both dPCR and NGS to detect HER2 amplification in the tumor tissues and blood of NSCLC patients. The ROC analysis results showed a good prediction result of dPCR in detecting HER2 amplification in blood samples. Furthermore, survival analysis showed that the median survival time was longer in the negative HER2 amplification group than in the positive group as detected by blood dPCR, which was accordant with the results of multivariate Cox regression. Finally, the amplification multiple in tissue samples was ≥1.59 times and amplification multiple in blood samples was ≥1.43 times the recommended cutoff values for dPCR detection in patients with NSCLC.
Amplification/overexpression of the HER2 gene is related to the occurrence and development of tumors (21-23). As for lung adenocarcinoma (LUAD), HER2 aberration could be detected in about 6% patients with no EFGR/KRAS/ALK alteration (24). In LUAD, patients with HER2 alteration would have higher risk of lung and bone metastases (25). Moreover, HER2 amplification was a mechanism of resistant to osimertinib (26). HER2 was also an actionable driver and currently some drugs targeted HER2 alteration had been tested in clinical trials (27,28). Based on these studies, detecting HER2 alterations could help us define a novel subset in EFGR/KRAS/ALK negative LUAD patients, evaluating the metastatic risk of patients, understanding EGFR-TKI resistance and selecting proper patients for HER2-TKI treatment. Thus, a suitable method for HER2 gene amplification/mutation detection is necessary for patient classification and cancer intervention (29). To date, the NGS method has been commonly used for HER2 amplification detection (30). A previous study indicated that NGS can identify the coexistence of HER2 amplification and mutation in NSCLC (31). However, NGS also has disadvantages; its application is limited by the reading length of 200–500 bp segments (32).
A recent cross-platform comparison study based on ctDNA samples obtained from patients with NSCLC showed that dPCR had a unique advantage in gene mutation detection compared with NGS (33). Researchers have shown the dPCR to be a more precise and less subjective alternative for quantifying HER2 DNA amplification in cancer (34). A previous study showed that dPCR was sensitive in detecting EGFR mutations from ctDNA in advanced NSCLC patients (14). The feasibility of dPCR for the quantitative and dynamic detection of EGFR mutations has been supported in comparison with NGS in lung cancer (35). Mehrotra et al. indicated that dPCR had 100% sensitivity in mutation detection and also revealed the correlation between OS and gene mutation (36). In our study, sensitivity and specificity analysis based on the ROC showed an excellent prediction result of dPCR detection on HER2 amplification. Meanwhile, the survival analysis showed that the median survival time between negative and positive HER2 DNA amplification groups detected by blood dPCR was significantly different, which was accordant with the result of multivariate Cox regression analysis. Thus, we speculated that dPCR could be applied in detecting HER2 amplification of NSCLC. And the HER2 amplification detected by dPCR also could be a potential method for predicting the OS of NSCLC patients.
The continuous research of targeted and immunotherapy drugs has resulted in the increase of treatment opportunities for NSCLC (37); however, it is challenging to collect tissue samples from lung cancer patients (38). The measurement of mutations in blood ctDNA may transform the management of cancer patient (39). It has been shown that dPCR is a useful method with high sensitivity and specificity for detecting EGFR mutation in plasma (40). A previous study showed that repeated measures on the same blood sample indicated that dPCR was less variable than another qPCR method (41). Li et al. indicated that dPCR improved EGFR mutation detection in the liquid but not tissue samples of NSCLC patients (42). In this study, we found that dPCR and NGS had identical efficiency in blood sample detection.
Meanwhile, sensitivity and specificity analysis based on the ROC showed that the P value in blood dPCR was below 0.05. This result demonstrated a good prediction result of dPCR detection on HER2 amplification in blood samples. Thus, we speculated that it was possible to use blood samples to detect HER2 amplification in NSCLC. However, there were still some limitations in the current study, due to its small sample size and retrospective study. Thus, further research based on a large sample size prospective study is needed to verify our findings.
In conclusion, dPCR detection of HER2 gene amplification might be a potential method to predict the OS of NSCLC. Blood samples could be used to detect HER2 amplification by dPCR.
Acknowledgments
The authors appreciate the academic support from AME Lung Cancer Collaborative Group.
Funding: This study was supported by the Projects Sponsored by the Development Fund of Shanghai Talents 2020.
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://dx.doi.org/10.21037/tlcr-21-860
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tlcr-21-860
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tlcr-21-860). HV and JX report that they are from Alphamab Oncology Ltd. JW and QZ report that they are from Genetron Health (Beijing) Co.,Ltd. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethic committee of Shanghai Pulmonary Hospital (No. K20-275) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Johnson BE. Divide and Conquer to Treat Lung Cancer. N Engl J Med 2016;375:1892-3. [Crossref] [PubMed]
- Chen Z, Fillmore CM, Hammerman PS, et al. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 2014;14:535-46. [Crossref] [PubMed]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013;63:11-30. [Crossref] [PubMed]
- Santa-Maria C, Jain S, Gradishar WJ. Human Epidermal Growth Factor Receptor 2 (HER2): Translating the Lab to the Clinic. In: Badve S, Gökmen-Polar Y. editors. Molecular Pathology of Breast Cancer. Cham: Springer, 2016:59-70.
- Minari R, Gnetti L, Lagrasta CA, et al. Emergence of a HER2-amplified clone during disease progression in an ALK-rearranged NSCLC patient treated with ALK-inhibitors: a case report. Transl Lung Cancer Res 2020;9:787-92. [Crossref] [PubMed]
- Heinmöller P, Gross C, Beyser K, et al. HER2 status in non-small cell lung cancer: results from patient screening for enrollment to a phase II study of herceptin. Clin Cancer Res 2003;9:5238-43. [PubMed]
- Garrido-Castro AC, Felip E. HER2 driven non-small cell lung cancer (NSCLC): potential therapeutic approaches. Transl Lung Cancer Res 2013;2:122-7. [PubMed]
- Matsuyama K, Oura S, Yoshimasu T, et al. The relationship between HER2/neu expression and drug resistance for non-small cell lung cancer. Lung Cancer 2000;29:13. [Crossref]
- Bartsch R, Berghoff A, Pluschnig U, et al. Impact of anti-HER2 therapy on overall survival in HER2-overexpressing breast cancer patients with brain metastases. Br J Cancer 2012;106:25-31. [Crossref] [PubMed]
- Sahoo S, Hwang H. Her2 FISH amplification in ER/PR/Her2 IHC negative breast cancer. Cancer Res 2015;abstr 75:P6-08-53.
- Parwani AV, Mohanty SK, Becich MJ. Pathology Reporting in the 21st Century: The Impact of Synoptic Reports and Digital Imaging. Lab Med 2008;39:582-6. [Crossref]
- Campomenosi P, Gini E, Noonan DM, et al. A comparison between quantitative PCR and droplet digital PCR technologies for circulating microRNA quantification in human lung cancer. BMC Biotechnol 2016;16:60. [Crossref] [PubMed]
- Cao L, Cui X, Hu J, et al. Advances in digital polymerase chain reaction (dPCR) and its emerging biomedical applications. Biosens Bioelectron 2017;90:459-74. [Crossref] [PubMed]
- Wang W, Song Z, Zhang Y. A Comparison of ddPCR and ARMS for detecting EGFR T790M status in ctDNA from advanced NSCLC patients with acquired EGFR-TKI resistance. Cancer Med 2017;6:154-62. [Crossref] [PubMed]
- Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol 2016;2:1014-22. [Crossref] [PubMed]
- Xu C, Zhang Y, Liu D, et al. P1.01-99 Detecting HER2 Alterations by Next Generation Sequencing (NGS) in Patients with Advanced NSCLC from the United States and China. J Thorac Oncol 2018;13:S502. [Crossref]
- Travis WD, Brambilla E, Burke AP, et al. Introduction to The 2015 World Health Organization Classification of Tumors of the Lung, Pleura, Thymus, and Heart. J Thorac Oncol 2015;10:1240-2. [Crossref] [PubMed]
- Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 2011;6:244-85. [Crossref] [PubMed]
- Liu Y, Yang M, Jiang T, et al. Quantitative Analysis of HER2 Amplification by Droplet Digital PCR in the Follow-Up of Gastric Cancer Patients Being Treated with Trastuzumab after Surgery. Gastroenterol Res Pract 2019;2019:1750329 [Crossref] [PubMed]
- Zhan Y, Zang H, Feng J, et al. Long non-coding RNAs associated with non-small cell lung cancer. Oncotarget 2017;8:69174-84. [Crossref] [PubMed]
- Afzal M, Amir M, Hassan MJ, et al. Clinical role of HER2 gene amplification and chromosome 17: a study on 154 IHC-equivocal cases of invasive breast carcinoma patients. Tumour Biol 2016;37:8665-72. [Crossref] [PubMed]
- Ercoli G, Lopez G, Ciapponi C, et al. Building Up a High-throughput Screening Platform to Assess the Heterogeneity of HER2 Gene Amplification in Breast Cancers. J Vis Exp 2017; [Crossref] [PubMed]
- Gutierrez C, Schiff R. HER2: biology, detection, and clinical implications. Arch Pathol Lab Med 2011;135:55-62. [Crossref] [PubMed]
- Arcila ME, Chaft JE, Nafa K, et al. Prevalence, clinicopathologic associations, and molecular spectrum of ERBB2 (HER2) tyrosine kinase mutations in lung adenocarcinomas. Clin Cancer Res 2012;18:4910-8. [Crossref] [PubMed]
- Patil T, Mushtaq R, Marsh S, et al. Clinicopathologic Characteristics, Treatment Outcomes, and Acquired Resistance Patterns of Atypical EGFR Mutations and HER2 Alterations in Stage IV Non-Small-Cell Lung Cancer. Clin Lung Cancer 2020;21:e191-204. [Crossref] [PubMed]
- Leonetti A, Sharma S, Minari R, et al. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer 2019;121:725-37. [Crossref] [PubMed]
- Hotta K, Aoe K, Kozuki T, et al. A Phase II Study of Trastuzumab Emtansine in HER2-Positive Non-Small Cell Lung Cancer. J Thorac Oncol 2018;13:273-9. [Crossref] [PubMed]
- Kris MG, Camidge DR, Giaccone G, et al. Targeting HER2 aberrations as actionable drivers in lung cancers: phase II trial of the pan-HER tyrosine kinase inhibitor dacomitinib in patients with HER2-mutant or amplified tumors. Ann Oncol 2015;26:1421-7. [Crossref] [PubMed]
- Mar N, Vredenburgh JJ. Dual HER2 Blockade in Non-Small Cell Lung Cancer Harboring a HER2 Mutation. Conn Med 2015;79:531-5. [PubMed]
- Holmes FA, Wang K, Levin MK, et al. Evidence of PIK3CA and TP53 co-mutation in breast cancer identification on next-generation sequencing (NGS) of ERBB2 (HER2)-amplified residual disease following preoperative anti-HER2 therapy. J Clin Oncol 2014;32:abstr 625.
- Chen R, Zhao J, Lin G, et al. MA 07.13 NGS Sequencing Based Liquid / Tissue Biopsy Identified Coexistence of HER2 Amplification and Mutation in Advanced NSCLC Patients. J Thorac Oncol 2017;12:S1830. [Crossref]
- Mileyko V, Ivanov M, Novikova E, et al. NGS for precision medicine in non-small cell lung cancer: Challenges and opportunities. Ann Oncol 2016;27:VI37. [Crossref]
- Garcia J, Forestier J, Dusserre E, et al. Cross-platform comparison for the detection of RAS mutations in cfDNA (ddPCR Biorad detection assay, BEAMing assay, and NGS strategy). Oncotarget 2018;9:21122-31. [Crossref] [PubMed]
- Belgrader P, Tanner SC, Regan JF, et al. Droplet digital PCR measurement of HER2 copy number alteration in formalin-fixed paraffin-embedded breast carcinoma tissue. Clin Chem 2013;59:991-4. [Crossref] [PubMed]
- Yang X, Zhuo M, Ye X, et al. Quantification of mutant alleles in circulating tumor DNA can predict survival in lung cancer. Oncotarget 2016;7:20810-24. [Crossref] [PubMed]
- Mehrotra M, Singh RR, Loghavi S, et al. Detection of somatic mutations in cell-free DNA in plasma and correlation with overall survival in patients with solid tumors. Oncotarget 2018;9:10259-71. [Crossref] [PubMed]
- Printz C. Immunotherapy drug improves survival of patients with squamous non-small cell lung cancer. Cancer 2015;121:3562-3. [Crossref] [PubMed]
- Al-Najjar H, Evison M, Crosbie PA, et al. Achieving a tissue diagnosis in PS3 patients with advanced lung cancer: what are the safety and diagnostic outcomes with EBUS? Lung Cancer 2015;88:236-7. [Crossref] [PubMed]
- Paredis D, Wildiers H, Vergote I, et al. A consecutive series of early breast cancers with a low estrogen receptor expression. Cancer Res 2017;77:abstr P6-09-12.
- Feng WN, Gu WQ, Zhao N, et al. Comparison of the SuperARMS and Droplet Digital PCR for Detecting EGFR Mutation in ctDNA From NSCLC Patients. Transl Oncol 2018;11:542-5. [Crossref] [PubMed]
- van Ginkel JH, van den Broek DA, van Kuik J, et al. Preanalytical blood sample workup for cell-free DNA analysis using Droplet Digital PCR for future molecular cancer diagnostics. Cancer Med 2017;6:2297-307. [Crossref] [PubMed]
- Li X, Liu Y, Shi W, et al. Droplet digital PCR improved the EGFR mutation diagnosis with pleural fluid samples in non-small-cell lung cancer patients. Clin Chim Acta 2017;471:177-84. [Crossref] [PubMed]


