
Development and validation of a nomogram for predicting survival in patients with surgically resected lung invasive mucinous adenocarcinoma
Introduction
Lung cancer is a prominent global health burden. Lung cancer causes the most cancer-related deaths worldwide (1), and adenocarcinoma is the most common histological type (2,3). Lung invasive mucinous adenocarcinoma (LIMA) is a variant of invasive lung adenocarcinoma, previously referred to as mucinous bronchioloalveolar carcinoma, according to the current 2015 World Health Organization (WHO) lung tumor classes (4).
LIMA consists of goblet and columnar cells with small basal-oriented nuclei, which are associated with abundant intracytoplasmic mucin. LIMAs are uncommon and only account for 2–10% of lung adenocarcinomas (5-8). Their relative rarity is one reason diagnosis of LIMAs is challenging. The non-mucinous lung adenocarcinoma features readily identifiable cell types, whereas the cytologic atypia of LIMA cells is usually inconspicuous. Additionally, LIMA often show lepidic-predominant growth, so it is more difficult to diagnose in the limited biopsy specimens and requires differentiation from metaplasia or bronchiolar adenoma. Moreover, the mixed mucinous adenocarcinoma may be difficult to diagnose from biopsy specimens, which depend on the biopsy site. In contrast, a mucinous adenocarcinoma can be easily identified by the presence of abundant mucin in the surgical specimen (9). However, the clinic-pathological characteristics of patients with LIMA are still unclear, due to the low incidence. Moreover, the prognosis of LIMAs remains controversial. Some studies reported that LIMAs might not be aggressive tumors, and the overall survival (OS) was significantly better than non-mucinous adenocarcinoma (10-12). Other reports showed conflicting results (5,13,14), suggesting that these entities tend to be very heterogeneous. Therefore, understanding the biology of LIMAs and identifying the high-risk factors for survival are crucial.
Nomograms quantify risk by revealing characteristics related to oncologic prognoses (15,16). Until now, no nomograms have been developed to forecast long-term survival outcomes after surgery in LIMAs. Consequently, our goal was to develop a novel nomogram for predicting the survival of surgically resected LIMAs using accepted clinic-pathological variables from the SEER database. A separate cohort was used to validate the nomogram externally.
This article is presented in accordance with the TRIPOD reporting checklist (available at https://dx.doi.org/10.21037/tlcr-21-562).
Methods
Study design and patients
We identified patients with LIMAs from 18 cancer registries within the population-based Surveillance, Epidemiology, and End Results (SEER) database [2008–2016] and the SEER*Stat program (version 8.3.5). The SEER database includes about 30% of the population of the United States of America.
In the SEER database, 1,792 patients conformed to the inclusion criteria of our study. All patients were diagnosed with invasive mucinous adenocarcinoma (8,480/3, 8,481/3, or 8,253/3) by histology (ICD-O-3) and underwent surgical resection. We excluded patients younger than 18 years, those with an overall survival of less than one month, and those missing essential information. Patients were randomized into training (n=1,254) and test cohorts (n=538) at a 7:3 ratio. Patients in the training cohort were retrospectively analyzed to construct the nomogram, which was internally validated by the test cohort. We also collected a data set (n=369) from the Cancer Hospital, Chinese Academy of Medical Sciences (CAMS, 2010–2017), to validate the nomogram externally. Inclusion and exclusion criteria were the same as the development cohort. If any information about predictors was missing, patients were excluded from the final risk model.
We collected the following patient-specific information from the databases: clinic-pathological characteristics (sex, age at diagnosis, marital status, primary site, race, tumor size, differentiation grade, tumor node metastasis (TNM) stage, visceral pleural invasion (VPI), and histological subtype); therapy details (extent of surgery, number of lymph nodes dissected, radiotherapy, and chemotherapy); and follow-up records (cause of death, vital survival status, and survival time). Two senior pathologists independently re-evaluated the histologic characteristics of the external validation cohort patients. Given that no well-established histological grading system with clearly defined criteria exists for LIMA, “differentiation grade” was determined according to the new histological grading paradigm for lung adenocarcinoma (8th edition TNM staging); this method is based on architectural predominant subtypes (I) G1 for lepidic adenocarcinoma, (II) G2 for acinar adenocarcinoma and papillary adenocarcinoma, and (III) G3 for micropapillary adenocarcinoma and solid adenocarcinoma.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (No. NCC1817) and individual consent for this retrospective analysis was waived.
Outcome
The predicted probability of 1-, 3-, and 5-year OS, based on baseline characteristics, was our primary outcome. OS was defined as the time between LIMA diagnosis and death (or last follow-up for cases where a death was not noted).
Statistical analysis
Nomogram development
We developed the nomogram using age, sex, histology, grade differentiation, primary site, laterality, TNM staging, VPI, radiotherapy, chemotherapy, and surgical extent and lymphadenectomy in the survival analysis. We explored the correlation between those variables and OS using the Kaplan-Meier method and the log-rank test. We developed a Cox proportional hazards model to evaluate independent prognostic factors, and those proposed factors were subsequently incorporated into a nomogram using the “survival” and “RMS” packages. We used the validation cohort to validate the model externally.
Assessing and comparing model performance
We verified the nomogram using calibration and discrimination curves. Calibration refers to the proximity between predicted probabilities and observed outcome frequencies. Discrimination refers to a nomogram’s ability to differentiate prognoses in patients with a concordance index (Harrell C-index) as quantization (17). The accuracy of the survival prediction for the maximum and final models were evaluated using time-dependent receiver operating characteristic (ROC) curves and area under the curves (AUCs) at 1, 3, and 5 years.
We performed a decision curve analysis (DCA) to compare the reliability of nomograms, which assessed whether alternative diagnostic or prognostic models were superior to others (18). We compared the nomogram to the traditional TNM staging system by C-index, determining net reclassification improvement (NRI) and integrated discrimination improvement (IDI) (19). The sum-score for each patient was determined using the points for each factor in the nomogram. The optimal cut-off value was determined using the “surv_cutpoint” function of the “survminer” package.
We classified patients into low- and high-risk groups according to each patient’s aggregate score in the nomogram. Based on the multivariable analyses, the selected independent risk factors were subjected to subgroup analysis. The OS for different groups were compared using the Kaplan-Meier method and log-rank test. We also assessed the effects of chemotherapy on OS during the subgroup analysis. An online version of the nomogram can be found at https://www.shinyapps.io/, to calculate predicted survival probability over time using survival-related factors.
Two-tailed statistical tests with a significance level set to P<0.05 were employed. The hazard ratio (HR) and 95% confidential interval (CI) for each variable with potential prognostic value was estimated using univariate and multivariate Cox proportional hazard regression models in a forward stepwise manner. Significant variables in univariate analyses (P<0.05) were carried into multivariate analyses. Statistical analyses were conducted with SPSS version 24.0 (IBM Corporation, Chicago, IL, USA) and R version 3.6.3. (http://www.r-project.org/).
Results
Participants
We identified 1,792 patients with LIMA from the SEER database, which were assigned randomly to either the training cohort (n=1,254) or the test cohort (n=538). Patient baseline characteristics are shown in Table 1. All characteristics among the patients were comparable in the different cohorts. The median patient ages in the training and test cohorts were 67 [interquartile range (IQR), 54–76] and 68.23 (IQR, 54–76) years, respectively. The male to female ratios were 545/709 and 219/319 in the training and validation cohorts, respectively. In the training cohort, 477 (52.6%) were stage I, 325 (35.8%) were stage II, 88 (9.7%) patients were stage III, and 17 (1.9%) patients were stage IV. Systematic mediastinal lymphadenectomy was performed in 1,103 (88.0%) patients, and 289 (23.0%) patients received adjuvant chemotherapy after surgery. In the test cohort, 335 (62.3%), 117 (21.7%), 63 (11.7%), and 23 (4.3%) patients were at stages I, II, III, and IV, respectively. Chemotherapy or radiotherapy was administered in 113 (21.0%) and 36 (6.7%) of patients. The median follow-up period was 44 months [95% confidence interval (CI): 42–46 months] and 42 months (95% CI: 39–46 months) in the training and test cohorts, respectively. The 5-year OS rates in the training and test cohorts were 17.15% and 15.24%.
Table 1
| Characteristics | Training cohort (n=1,254), n (%) | Test cohort (n=538), n (%) | External validation cohort (n=369), n (%) |
|---|---|---|---|
| Age (mean ± SD, years) | 67.75±10.62 | 68.23±10.54 | 57.77±11.04 |
| Sex | |||
| Male | 545 (43.5) | 219 (40.7) | 217 (58.8) |
| Female | 709 (56.5) | 319 (59.3) | 152 (41.2) |
| Race | |||
| White | 1,058 (84.4) | 448 (83.3) | 0 |
| Others | 196 (15.6) | 90 (16.7) | 369 (100.0) |
| Primary site | |||
| Upper lobe | 484 (38.6) | 198 (36.8) | 122 (33.1) |
| Middle lobe | 74 (5.9) | 30 (5.6) | 19 (5.1) |
| Lower lobe | 683 (54.5) | 303 (56.3) | 206 (55.8) |
| Others | 13 (1.0) | 7 (1.3) | 22 (6.0) |
| Laterality | |||
| Right | 705 (56.2) | 317 (58.9) | 196 (53.1) |
| Left | 549 (43.8) | 221 (41.1) | 173 (46.9) |
| VPI | |||
| No/unknown | 1,147 (91.5) | 485 (90.1) | 194 (52.6) |
| Yes | 107 (8.5) | 53 (9.9) | 175 (47.4) |
| Differentiation grade | |||
| I | 701 (55.9) | 283 (52.6) | 79 (21.4) |
| II | 463 (36.9) | 210 (39.0) | 181 (49.1) |
| III/IV | 90 (7.2) | 45 (8.4) | 109 (29.5) |
| Histological subtype | |||
| Pure LMIA | 1128 (90.0) | 488 (90.7) | 220 (59.6) |
| Mixed LMIA | 126 (10.0) | 50 (9.3) | 149 (40.4) |
| Extent of surgery | |||
| Lobectomya | 907 (72.3) | 369 (68.6) | 315 (85.4) |
| Sub-lobectomy | 296 (23.6) | 144 (26.8) | 50 (13.6) |
| Pneumonectomyb | 51 (4.1) | 25 (4.6) | 4 (1.1) |
| Tumor size (mean ± SD, mm) | 44.66±98.10 | 47.63±113.00 | 33.28±23.12 |
| T stage | |||
| T1 | 548 (43.7) | 251 (46.7) | 126 (34.1) |
| T2 | 409 (32.6) | 164 (30.5) | 195 (52.8) |
| T3 | 238 (19.0) | 100 (18.6) | 39 (10.6) |
| T4 | 59 (4.7) | 23 (4.3) | 9 (2.4) |
| N stage | |||
| N0 | 1,067 (85.1) | 515 (95.7) | 276 (74.8) |
| N+ | 167 (14.9) | 23 (4.3) | 93 (25.2) |
| M stage | |||
| M0 | 1,194 (95.2) | 515 (95.7) | 361 (97.8) |
| M1 | 60 (4.8) | 23 (4.3) | 8 (2.2) |
| Number of LND (mean ± SD) | 9.12±9.98 | 9.30±9.63 | 17.32±10.61 |
| Adjuvant radiotherapy | |||
| Presence | 97 (7.7) | 36 (6.7) | 21 (5.69) |
| Absence | 1,157 (92.3) | 502 (93.3) | 348 (94.31) |
| Adjuvant chemotherapy | |||
| Presence | 289 (23.0) | 113 (21.0) | 126 (34.15) |
| Absence | 965 (77.0) | 425 (79.0) | 243 (65.85) |
a, including lobectomy and bilobectomy; b, including pneumonectomy and lobectomy extended. SD, standard deviation; LIMA, lung mucinous invasive adenocarcinoma; VPI, visceral pleural invasion; LND, lymph nodes dissected.
Univariate and multivariate Cox proportional hazard analyses
Univariate and multivariate Cox regression analyses were employed to detect factors that significantly correlate with OS, as shown in Table 2. The univariate analysis revealed that older age (P<0.0001), sex (P<0.0001), race (P=0.023), marital status (P=0.036), larger tumor size (P<0.0001), primary site (P=0.004), poorly differentiated status (P<0.0001), more advanced T, N, and M stages (all P<0.0001), the extent of surgery (P<0.0001), lymphadenectomy (P<0.0001), and the absence of radiotherapy or chemotherapy (both P<0.0001) significantly correlated with shorter OS. The variables identified in the univariate analyses were employed in the multivariate analysis to determine if they independently associated with OS. Multivariate analyses showed that age (P<0.0001), sex (P=0.001), differentiation grade (P<0.0001), T stage (P<0.0001), N stage (P<0.0001), M stage (P=0.001), the extent of surgery (P<0.0001), and lymphadenectomy (P=0.039) were able to predict the OS of LIMA patients.
Table 2
| Variable | Univariable analysis | Multivariable analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | ||
| Age (years) | |||||||
| <60 | Reference | Reference | |||||
| ≥60 | 1.824 | 1.361–2.445 | <0.0001 | 1.861 | 1.375–2.519 | <0.0001 | |
| Sex | |||||||
| Female | Reference | Reference | |||||
| Male | 1.543 | 1.261–1.888 | <0.0001 | 1.432 | 1.158–1.773 | 0.001 | |
| Race | |||||||
| White | Reference | Reference | |||||
| Others | 0.692 | 0.504–0.950 | 0.023 | 0.787 | 0.571–1.086 | 0.145 | |
| Martial status | |||||||
| Married | Reference | Reference | |||||
| Others | 1.251 | 1.015–1.541 | 0.036 | 1.064 | 0.857–1.320 | 0.574 | |
| Tumor size (mm) | |||||||
| <50 | Reference | Reference | |||||
| ≥50 | 1.527 | 1.210–1.929 | <0.0001 | 1.137 | 0.856–1.510 | 0.376 | |
| Primary site | 0.004 | 0.062 | |||||
| Lower lobe | Reference | Reference | |||||
| Middle lobe | 0.983 | 0.627–1.542 | 0.942 | 0.943 | 0.594–1.496 | 0.803 | |
| Upper lobe | 1.090 | 0.881–1.347 | 0.427 | 0.977 | 0.783–1.220 | 0.840 | |
| Others | 3.422 | 1.754–6.676 | <0.0001 | 2.515 | 1.264–5.005 | 0.009 | |
| Laterality | |||||||
| Right | Reference | ||||||
| Left | 1.184 | 0.967–1.449 | 0.102 | ||||
| Histological subtype | |||||||
| Pure LMIA | Reference | ||||||
| Mixed LMIA | 1.208 | 0.892–1.635 | 0.222 | ||||
| VPI | |||||||
| No/unknown | Reference | ||||||
| Yes | 1.211 | 0.859–1.708 | 0.274 | ||||
| Differentiation grade | <0.0001 | <0.0001 | |||||
| I | Reference | Reference | |||||
| II | 1.940 | 1.566–2.404 | <0.0001 | 1.665 | 1.326–2.091 | <0.0001 | |
| III/IV | 2.889 | 2.054–4.064 | <0.0001 | 1.947 | 1.339–2.832 | <0.0001 | |
| Extent of surgery | <0.0001 | <0.0001 | |||||
| Sub-lobectomy | Reference | Reference | |||||
| Lobectomya | 0.551 | 0.443–0.686 | <0.0001 | 0.579 | 0.434–0.773 | <0.0001 | |
| Pneumonectomyb | 0.709 | 0.426–1.178 | 0.184 | 0.453 | 0.259–0.791 | 0.005 | |
| Lymphadenectomy | |||||||
| No | Reference | Reference | |||||
| Yes | 0.518 | 0.399–0.674 | <0.0001 | 0.701 | 0.501–0.982 | 0.039 | |
| T stage | <0.0001 | <0.0001 | |||||
| T1 | Reference | Reference | |||||
| T2 | 1.595 | 1.239–2.053 | <0.0001 | 1.549 | 1.182–2.029 | 0.002 | |
| T3 | 2.204 | 1.681–2.890 | <0.0001 | 2.153 | 1.544–3.004 | <0.0001 | |
| T4 | 6.148 | 4.279–8.834 | <0.0001 | 2.167 | 1.358–3.458 | 0.001 | |
| N stage | |||||||
| N0 | Reference | Reference | |||||
| N+ | 2.091 | 1.843–2.372 | <0.0001 | 1.645 | 1.379–1.961 | <0.0001 | |
| M stage | |||||||
| M0 | Reference | Reference | |||||
| M1 | 3.557 | 2.571–4.922 | <0.0001 | 1.975 | 1.343–2.906 | 0.001 | |
| Radiotherapy | |||||||
| Presence | Reference | Reference | |||||
| Absence | 2.750 | 2.078–3.639 | <0.0001 | 1.281 | 0.925–1.775 | 0.136 | |
| Chemotherapy | |||||||
| Presence | Reference | Reference | |||||
| Absence | 2.122 | 1.718–2.622 | <0.0001 | 0.889 | 0.673–1.173 | 0.405 | |
a, including lobectomy and bilobectomy; b, including pneumonectomy and lobectomy extended. HR, hazard ratio; CI, confidence interval; MIA, mucinous invasive adenocarcinoma; VPI, visceral pleural invasion.
Development of nomogram for OS
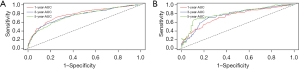
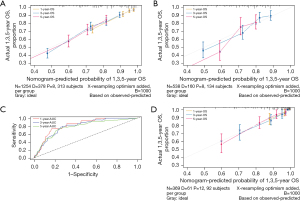
A nomogram was constructed for predicting 1-, 3-, and 5-year OS using the independent factors identified in the training cohort, including age, gender, differentiation grade, T, N, and M stage, the extent of surgery, and lymphadenectomy (Figure 1). The probability of individual survival was determined by adding up all scores for each factor. The nomogram indicated that the N stage was the most crucial factor affecting the outcome, followed by the T stage, differentiation grade, M stage, the extent of surgery, age, lymphadenectomy, and sex. The C-indices in the training and test cohorts were 0.735 (95% CI: 0.728–0.742) and 0.736 (95% CI: 0.7255–0.7465) for OS. Based on the ROC curve analyses for the 1-, 3-, and 5-year OS, the AUC values were 0.782, 0.756, and 0.747, respectively, in the training cohort (Figure 2A) and 0.738, 0.756, and 0.798, respectively, in the test cohort (Figure 2B). The C-indices and AUC values indicated that the nomogram possessed accurate predictive and adequate discriminative abilities. Furthermore, we generated calibration plots for probable 1-, 3-, and 5-year OS. All generated plots closely approximated the observed estimates in the training and test cohorts (Figure 3A,3B), demonstrating that the two groups were well-calibrated. The newly developed nomogram reliably predicted survival in patients with LIMA after surgical resection.
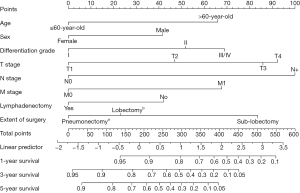
Validation of the nomogram
The external validation cohort included 369 patients in the final analysis. Table 1 shows the baseline characteristics of the external cohort. Solitary-type was found in 348 (94.31%) patients, while pneumonic-type was found in 21 (5.69%) patients. The follow-up time was 53.6 months (median, 95% CI: 50.2–58.0 months) and the 5-year OS rate was 34.69%. The nomogram C-index for predicting OS was 0.773 (95% CI: 0.760–0.786). AUC values for the 1-, 3-, and 5-year OS were 0.826, 0.800, and 0.777, respectively (Figure 3C). Moreover, the external calibration plots for predicted probabilities of 1-, 3-, and 5-year OS also showed adequate consistency (Figure 3D).
The prognostic nomogram in the clinical practice
An exploratory examination of the nomogram predictive value for risk stratification was conducted. A total risk score was determined for each patient in the training cohort. Based on these calculations, the cut-off value of the sum-score was 263. We then grouped the training cohort into high-risk (scores >263) or low-risk (scores ≤263) groups (Figure S1). The low-risk patients had significantly longer OS compared with high-risk patients (log-rank, P<0.001) (Figure 4A). The same cut-off value for OS also differentiated high- and low-risk groups in the test cohort (log-rank, P<0.001) and external validation cohort (log-rank, P<0.001) (Figure 4B,4C). Similarly, the high-risk group had a significantly shorter OS than the low-risk group in the three integrated cohorts (log-rank, P<0.001) (Figure 4D).
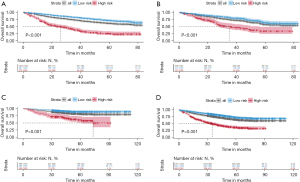
To further verify the nomogram’s stability in different clinical subgroups, we conducted a stratified analysis in patients with pure LIMA and mixed LIMA to evaluate the nomogram’s predictive ability. The nomogram could accurately classify OS for patients in high- and low-risk groups (log-rank, all P<0.001) (Figure S2).
Comparison of predictive accuracy for OS in the current nomogram versus the conventional AJCC staging systems
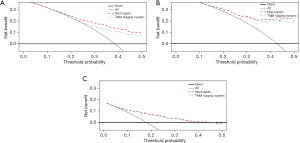
The clinical validity of the nomogram was verified using a DCA, demonstrating the superior benefits of the new model compared with the 7th edition TNM staging system. The nomogram was better than the conventional AJCC staging system at predicting survival in LIMA patients (Figure 5A-5C). The C-index for OS of our nomogram was significant higher than that of the conventional TNM staging system in the training cohort (0.735 vs. 0.694, respectively), in the test cohort (0.736 vs. 0.712, respectively), and the external validation cohort (0.773 vs. 0.741, respectively). Notably, the NRI values were 0.233 (95% CI: 0.128 to 0.384), 0.084 (95% CI: −0.015 to 0.288), and 0.245 (95% CI: −0.106 to 0.409) in the training, test, and external validation cohorts, respectively. The IDI values were 0.245 (95% CI: −0.106 to 0.409), 0.038 (95% CI: 0.014–0.087), and 0.032 (95% CI: 0.010–0.113) in the training, test, and external validation cohorts, respectively. Thus, our nomogram predicted prognosis better than conventional TNM staging.
Development of a web server for accessing the new model
An online version of the nomogram is located at https://doctor1.shinyapps.io/DynNomap/ (Figure S3) and may help guide risk-reducing interventions and survival predictions for clinicians. Predicted survival can be determined by inputting clinical features into the web server.
Discussion
LIMA is a distinct pulmonary malignancy, whose biological behaviors and clinicopathological features remain unclear. Several previous studies reported that the clinicopathological characteristics and prognosis of LIMA are unique from other subtypes of lung adenocarcinoma (20-23). However, survival data for LIMA in the few published reports tend to be contradictory. These differences suggested that the prognosis of LIMA patients is heterogeneous. Given that the previous LIMA studies were limited to small sample sizes, clarification of the prognostic factors based on a large cohort and development of a model to predict LIMA patient overall survival is urgently needed.
In the present study, we enrolled 2,161 patients with LIMA, a much bigger cohort than previous LIMA studies. From the SEER database, 1,792 patients with LIMA were identified, which were split randomly into training (n=1,254) and test cohorts (n=538). We used univariate and multivariate Cox proportional hazard analyses to analyze independent prognostic factors based on clinicopathological characteristics in the training cohort. We then identified seven independent prognostic factors, including age, sex, differentiation grade, the extent of surgery, lymphadenectomy, and T, N, and M stages, which were incorporated into the nomogram. The nomogram was successfully validated using an external cohort of 369 patients with LIMA. Encouragingly, the proposed nomogram revealed excellent discrimination and calibration. This is the first nomogram able to predict OS in patients with surgically-resected LIMA, based on the SEER database and an external validation cohort. More importantly, the number of patients included in most previous studies was relatively small. In contrast, at present, our study is the largest cohort of LIMA patients with long-term follow-up.
We identified differentiation grade, extent of surgery, and lymphadenectomy as independent predictors for overall survival, in addition to the risk factors of age, sex, and T, N, and M stages. Previous studies indicated that several of these variables were risk factors related to survival in patients with LIMA (10,24). In this study, female patients have a better survival than male patients, which may be related to the more populations of stage I in female patients. Moreover, female patients are more likely to be younger, never-smokers, and demonstrate EGFR-driver mutations and the p53 wild-type transcription factor—all of which are protective factors in lung cancer (25,26). Of note, surgical methods and regional lymph node examination are independent prognostic factors for LIMAs, as demonstrated by this study. Surgical resection is the primary treatment for operable LIMAs, and most guidelines indicate that lobectomy is the most common surgery for patients with lung adenocarcinoma. Interestingly, we found that patients who underwent pneumonectomy or lobectomy had a better prognosis than those who underwent sub-lobectomy. In the prognostic nomogram, the risk score of pneumonectomy was less than lobectomy (0 vs. 23 points); however, there was no significant difference between pneumonectomy and lobectomy (P=0.322). Although resection of more tissue may improve survival for patients with pneumonic-type LIMA, lobectomy is still gold-standard surgical procedure for LIMA. Furthermore, patients who had a lymphadenectomy had a more favorable survival than those who did not. These results are consistent with Moon et al. (11), suggesting that patients undergoing lymphadenectomy with adequate pathologic staging had improved survival.
Our univariate analysis results indicate that adjuvant chemotherapy and radiotherapy are significantly related to prolonged OS. However, these same parameters were negatively associated with OS in the multivariate analysis. This may be attributed to interactions between chemotherapy and age and TNM stage. Luo et al. found no significant survival benefits from adjuvant chemotherapy for postoperative patients with LIMA (8). Cha et al. reported that in stage IV LIMA patients, treatment with chemotherapy did not improve OS (12). Moreover, LIMA patients possessed a distinct molecular signature and lack of targetable mutations (20,22,27,28). Thus, this population might not benefit from targeted therapy with tyrosine kinase receptor inhibitors, and a new therapeutic approach is urgently warranted.
In the present study, 3.33% of lung cancer patients in the SEER database had LIMA and 2.13% of lung cancer patients in the Cancer Hospital, Chinese Academy of Medical Sciences had LIMA. The most common primary site of LIMA was the lower lobe. LIMA patients tended to be less advanced in the SEER database and validation cohort, consistent with previous results (10-12). Given that there were some differences between pure LIMA and mixed LIMA, we conducted a subgroup analysis of OS. This analysis demonstrated that the high-risk group had a significantly shorter OS compared with the low-risk group in both the pure and mixed LIMA subgroups. Of note, we used the nomogram to stratify patients with LIMA into prognostic risk groups. There is an urgent need to identify appropriate candidates for postoperative adjuvant therapy.
The risk factors incorporated into the survival prediction model were readily ascertainable clinicopathological variables. Their inclusion minimizes patient data collection variability and improves the general applicability of this model, making it an easy-to-use scoring system. A few nomograms have been developed to predict the prognosis of patients with mucinous adenocarcinoma. Chen et al. developed a prognostic nomogram for LIMA with a C-index of 0.724 (10). However, all LIMA patients were enrolled, including the advanced stage patients diagnosed with a transbronchial biopsy. We focused only on LIMA patients who underwent surgery because diagnosis of LIMA based on surgical specimens can be easily identified while its definitive diagnosis by transbronchial biopsy before surgery is more challenging.
The American Joint Committee on Cancer TNM classification is the most extensively utilized staging system (29); however, patient survival at the same stage varies widely (30-32). Other independent prognostic factors, including sex, age, histology, and treatment-related factors, may contribute significantly to predicting individualized survival. Therefore, we aimed to develop a model to better stratify LIMAs into different prognostic categories. Our study revealed that the C-index for 1-/3-/5-year OS in the nomogram was higher compared with the traditional TNM staging system and previously published model. The nomogram displayed adequate calibration plots for the 1-/3-/5-year OS scores in the training, test, and external validation cohorts. DCA is a novel method for assessing diagnostic tests and predictive models. In this study, the clinical utility was in the proper range. Additionally, the ideal net benefits (the C-index, NRI and IDI values) of the new model were superior to the 7th edition TNM staging system, suggesting that this model may be a practical instrument to provide personalized predictions for patients with LIMA.
Although the nomogram proved to be an effective independent prognostic determiner for patients with LIMA, there are several limitations. First, this was a retrospective study, incurring inevitable bias. Further prospective studies may be required to validate the risk model. Second, the nomogram did not include some crucial predictors, such as a history of smoking, lymphovascular invasion, chemotherapy regimens, and the predominant growth pattern of LIMAs, due to the non-availability of this information in the SEER database. Third, potential interaction terms were not considered in this nomogram for improving its convenience and interpretability. Consideration of all reasonable interaction terms in the model construction may result in a better survival prediction; however, it would make the model more complex and difficult to use for clinical practice. Last but not least, the lung cancer TNM staging system applied in our study was the 7th edition, because the current 8th edition stage information was not available in the SEER database. Although there are some discrepancies between different editions of staging systems, the prognostic values are consistent and they are constantly being optimized. Despite these limitations, our prognostic nomogram, combined with the online web server, is an essential and useful model, which provides accurate and individualized survival predictions in LIMA patients.
Conclusions
We developed and validated the first nomogram for patients with surgically resected LIMA based on the SEER database and a large external cohort. This nomogram can improve prognostic assessment, aid individual treatment, and guide follow-up management strategies for patients with LIMA.
Acknowledgments
All authors would like to thank all patients and staff who have participated in and contributed to the SEER program tumor registries. We also would like to thank Karen Uray and Erin P. Silverman for their help in polishing our paper.
Funding: This work was supported by funds from the National Key Research and Development Program of China (2018YFC1311601, 2016YFC1303200).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://dx.doi.org/10.21037/tlcr-21-562
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tlcr-21-562
Peer Review File: Available at https://dx.doi.org/10.21037/tlcr-21-562
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tlcr-21-562). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (No. NCC1817) and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Gridelli C, Rossi A, Carbone DP, et al. Non-small-cell lung cancer. Nat Rev Dis Primers 2015;1:15009. [Crossref] [PubMed]
- Duma N, Santana-Davila R, Molina JR. Non-Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin Proc 2019;94:1623-40. [Crossref] [PubMed]
- Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. [Crossref] [PubMed]
- Yoshizawa A, Motoi N, Riely GJ, et al. Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage I cases. Mod Pathol 2011;24:653-64. [Crossref] [PubMed]
- Sun F, Wang P, Zheng Y, et al. Diagnosis, clinicopathological characteristics and prognosis of pulmonary mucinous adenocarcinoma. Oncol Lett 2018;15:489-94. [PubMed]
- Cai D, Li H, Wang R, et al. Comparison of clinical features, molecular alterations, and prognosis in morphological subgroups of lung invasive mucinous adenocarcinoma. Onco Targets Ther 2014;7:2127-32. [PubMed]
- Luo J, Wang R, Han B, et al. Analysis of the clinicopathologic characteristics and prognostic of stage I invasive mucinous adenocarcinoma. J Cancer Res Clin Oncol 2016;142:1837-45. [Crossref] [PubMed]
- Cha YJ, Shim HS. Biology of invasive mucinous adenocarcinoma of the lung. Transl Lung Cancer Res 2017;6:508-12. [Crossref] [PubMed]
- Chen Z, Li M, Ma K, et al. Analysis of the clinicopathological characteristics, genetic phenotypes, and prognostic of pure mucinous adenocarcinoma. Cancer Med 2020;9:517-29. [Crossref] [PubMed]
- Moon SW, Choi SY, Moon MH. Effect of invasive mucinous adenocarcinoma on lung cancer-specific survival after surgical resection: a population-based study. J Thorac Dis 2018;10:3595-608. [Crossref] [PubMed]
- Cha YJ, Kim HR, Lee HJ, et al. Clinical course of stage IV invasive mucinous adenocarcinoma of the lung. Lung Cancer 2016;102:82-8. [Crossref] [PubMed]
- Casali C, Rossi G, Marchioni A, et al. A single institution-based retrospective study of surgically treated bronchioloalveolar adenocarcinoma of the lung: clinicopathologic analysis, molecular features, and possible pitfalls in routine practice. J Thorac Oncol 2010;5:830-6. [Crossref] [PubMed]
- Yanagawa N, Shiono S, Abiko M, et al. New IASLC/ATS/ERS classification and invasive tumor size are predictive of disease recurrence in stage I lung adenocarcinoma. J Thorac Oncol 2013;8:612-8. [Crossref] [PubMed]
- Iasonos A, Schrag D, Raj GV, et al. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol 2008;26:1364-70. [Crossref] [PubMed]
- Liang W, Zhang L, Jiang G, et al. Development and validation of a nomogram for predicting survival in patients with resected non-small-cell lung cancer. J Clin Oncol 2015;33:861-9. [Crossref] [PubMed]
- Harrell FE Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med 1996;15:361-87. [Crossref] [PubMed]
- Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making 2006;26:565-74. [Crossref] [PubMed]
- Van Calster B, Van Huffel S. Integrated discrimination improvement and probability-sensitive AUC variants. Stat Med 2010;29:318-9. [Crossref] [PubMed]
- Nakagomi T, Goto T, Hirotsu Y, et al. Genomic Characteristics of Invasive Mucinous Adenocarcinomas of the Lung and Potential Therapeutic Targets of B7-H3. Cancers (Basel) 2018;10:478. [Crossref] [PubMed]
- Lee HY, Cha MJ, Lee KS, et al. Prognosis in Resected Invasive Mucinous Adenocarcinomas of the Lung: Related Factors and Comparison with Resected Nonmucinous Adenocarcinomas. J Thorac Oncol 2016;11:1064-73. [Crossref] [PubMed]
- Shim HS, Kenudson M, Zheng Z, et al. Unique Genetic and Survival Characteristics of Invasive Mucinous Adenocarcinoma of the Lung. J Thorac Oncol 2015;10:1156-62. [Crossref] [PubMed]
- Ritterhouse LL, Vivero M, Mino-Kenudson M, et al. GNAS mutations in primary mucinous and non-mucinous lung adenocarcinomas. Mod Pathol 2017;30:1720-7. [Crossref] [PubMed]
- Geles A, Gruber-Moesenbacher U, Quehenberger F, et al. Pulmonary mucinous adenocarcinomas: architectural patterns in correlation with genetic changes, prognosis and survival. Virchows Arch 2015;467:675-86. [Crossref] [PubMed]
- Chapman AM, Sun KY, Ruestow P, et al. Lung cancer mutation profile of EGFR, ALK, and KRAS: Meta-analysis and comparison of never and ever smokers. Lung Cancer 2016;102:122-34. [Crossref] [PubMed]
- Haupt S, Caramia F, Herschtal A, et al. Identification of cancer sex-disparity in the functional integrity of p53 and its X chromosome network. Nat Commun 2019;10:5385. [Crossref] [PubMed]
- Righi L, Vatrano S, Di Nicolantonio F, et al. Retrospective Multicenter Study Investigating the Role of Targeted Next-Generation Sequencing of Selected Cancer Genes in Mucinous Adenocarcinoma of the Lung. J Thorac Oncol 2016;11:504-15. [Crossref] [PubMed]
- Hwang DH, Sholl LM, Rojas-Rudilla V, et al. KRAS and NKX2-1 Mutations in Invasive Mucinous Adenocarcinoma of the Lung. J Thorac Oncol 2016;11:496-503. [Crossref] [PubMed]
- Goldstraw P, Chansky K, Crowley J, et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2016;11:39-51. [Crossref] [PubMed]
- Zeng Y, Mayne N, Yang CJ, et al. A Nomogram for Predicting Cancer-Specific Survival of TNM 8th Edition Stage I Non-small-cell Lung Cancer. Ann Surg Oncol 2019;26:2053-62.
- Shang X, Yu H, Lin J, et al. A Novel Nomogram including AJCC Stages Could Better Predict Survival for NSCLC Patients Who Underwent Surgery: A Large Population-Based Study. J Oncol 2020;2020:7863984. [Crossref] [PubMed]
- Sculier JP, Chansky K, Crowley JJ, et al. The impact of additional prognostic factors on survival and their relationship with the anatomical extent of disease expressed by the 6th Edition of the TNM Classification of Malignant Tumors and the proposals for the 7th Edition. J Thorac Oncol 2008;3:457-66.


