
The miR-4306/IGF2R axis modulates the lung adenocarcinoma response to irradiation in vitro and in vivo
Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide (1). Non-small-cell lung cancer (NSCLC) represent approximately 80% to 85% of all lung carcinomas, and in the recent years, adenocarcinoma histological type has displaced squamous cell carcinoma worldwide as the most common form of NSCLC accounting for more than 50% of all primary lung cancers (2). Although multiple treatment modalities for lung adenocarcinoma have been proposed, patients’ prognosis remains unsatisfactory, especially in stages III–IV NSCLC (3). Moreover, radiotherapy is an integral part of treatment for lung adenocarcinoma and is considered as a curative treatment for stage I NSCLC (4). Lung adenocarcinoma presents substantial heterogeneity in radiation sensitivity. Therefore, discovery of novel precise biomarkers for prognosis and radiation sensitivity is warranted. Furthermore, a better understanding of molecular mechanisms underlying tumor progression and radiation resistance may promote the development of therapeutic strategies for lung adenocarcinoma.
MicroRNAs (miRNAs), a class of single-stranded non-coding RNAs of approximately 22 nucleotides, are potent regulators of gene expression through mRNA decay and translational inhibition (5). miRNAs act as oncogenes or tumor suppressor genes, which play critical roles on tumorigenesis and cancer progression of multiple cancers such as lung cancer, glioblastoma and gastric cancer (6-8). In addition, miRNAs have been reported to be involved in the radiosensitivity of different cancer cell types. For example, miR-622 down-regulates Rb expression, thus reducing radioresistance in colorectal cancer (9). Qu et al. (10) found that miR-205 could directly target PTEN (phosphatase and tensin homolog deleted on chromosome 10) to participate in the radioresistance of nasopharyngeal carcinoma. As recently reported, the radioresistance of lung adenocarcinoma is associated with dysregulation of miR-451/c-Myc-survivin/rad-51 signaling. Furthermore, miR-451 could directly target c-Myc to enhance the radioresistance of cancer cell apoptosis (11). With the development of high-throughput sequencing technology, an increasing number of differentially expressed miRNAs have been identified and may reveal promising biomarkers and therapeutic targets such as in head neck cancers (12). By analyzing online data sets from Gene Expression Omnibus (GEO) and The Cancer Genome Atlas Lung Adenocarcinoma (TCGA-LUAD), we discovered miR-4306 overexpression to be significantly related to poor prognosis of lung adenocarcinoma. And the expression level of miR-4306 was increased in lung cancerous tissue samples according to GSE135918 and TCGA-LUAD databases. Nevertheless, the specific roles of miR-4306 in lung adenocarcinoma progression has not been fully reported and need further clarification.
Insulin like growth factor 2 receptor (IGF2R), as a targeted gene of miR-4306 predicted by bioinformatics analysis in our research, encodes a receptor for both insulin-like growth factor 2 and mannose 6-phosphate. This receptor has various functions, including in the intracellular trafficking of lysosomal enzymes, the activation of transforming growth factor beta, and the degradation of insulin-like growth factor 2 (13). A lot of research had also disclosed that IGF2R involved in multiple malignant tumors, including cervical cancer (14), bladder cancer (15), osteosarcoma (16) and so on. However, the effective mechanism by which IGF2R exhibits its function in lung adenocarcinoma, especially its reciprocities with non-coding RNA, still remains vague.
In the present research, for the first time, we tried to reveal the biological functions and the potential molecular mechanism of miR-4306 in lung adenocarcinoma progression and radiation resistance by in vivo and in vitro experiments. We observed that overexpression of miR-4306 promoted cell viability and invasive abilities and attenuated the inhibitory effect of irradiation on malignant cancer cell behaviors by restraining IGF2R expression level. Our basic experimental outcomes may provide sufficient results to uncover a newfound miRNA/mRNA axis of miR-4306/IGF2R in lung adenocarcinoma progression and radiation resistance, supplying a crucial perception concerning the regulatory theory of miRNA in lung adenocarcinoma advancement and new therapeutic options of neoplastic diseases.
We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/tlcr-21-890).
Methods
Clinical sampling
Twenty-seven human lung adenocarcinoma specimens and 27 adjacent non-tumor lung tissue specimens were collected from patients who underwent surgery from 2019 to 2020 at Beijing Hospital, National Center of Gerontology, Institute of Geriatric Medicine, Chinese Academy of Medical Sciences. None of the patients included in this study underwent any preoperative radiation or chemotherapy. An experienced pathologist evaluated the pathological stage, grade, and nodal status. All procedures performed in this study involving human participants were approved by Research Ethics Committee of the Beijing Hospital, National Center of Gerontology, Institute of Geriatric Medicine, Chinese Academy of Medical Sciences (approval number: 2018BJYYEC-221-01) and were conducted in accordance with the Helsinki declaration (as revised in 2013). Written informed consent was obtained from all patients.
Cell lineage and cell culture
The normal human lung epithelial cell line BEAS-2B (CRL-960) was obtained from ATCC (Manassas, VA, USA) and cultured using a BEGM Kit (Catalog No. CC-3170; Lonza, Basel, Switzerland). The lung adenocarcinoma cell line A549 (CCL-185) was obtained from ATCC and cultured in F-12K medium (ATCC) supplemented with 10% FBS (Invitrogen, Carlsbad, CA, USA). The lung adenocarcinoma cell line 95-D (3131C0001000700061) was obtained from the Cell Resource Center, Shanghai Institute of Life Sciences, Chinese Academy of Sciences (Shanghai, China) and cultured in RPMI 1640 medium (Gibco, Waltham, MA, USA) supplemented with 10% FBS (Invitrogen).
X-ray irradiation
An X-ray generator (TITAN, Shimazu Co., Kyoto, Japan) set at 200 kVp and 20 mA was used for irradiation. Irradiation occurred through a copper and aluminum filter thickness of 0.5 mm, producing an effective energy of approximately 83 keV. The samples received X-ray doses at a dose rate of approximately 1 Gy/min. The doses used for cell line irradiation were 0, 2, 4, 6, 8, and 10 Gy with one fraction.
Cell transfection
miR-4306 overexpression and inhibition were achieved in target cells by transfecting agomir-4306 or antagomir-4306 (GenePharma, Shanghai, China). IGF2R overexpression was achieved by transfecting an IGF2R-overexpressing plasmid based on the pLVX-puro plasmid (Catalog No. 632164; Takara Bio, Mountain View, CA, USA). IGF2R knockdown was achieved by transfecting small interfering RNA against IGF2R (si-IGF2R#1/2/3, GenePharma). The specific sequences are listed in Table 1. All transfection was achieved using Lipofectamine 3000 (Invitrogen) reagents.
Table 1
| Name | Forward | Reverse |
|---|---|---|
| RT-PCR miR-4529-3p | RT: GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACACGGGC F: GCCATTGGACTGCTGATG |
R: CAGTGCGTGTCGTGGA |
| RT-PCR miR-4306 | RT: GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACTACTGC F: GCCGCTGGAGAGAAAG |
R: CAGTGCGTGTCGTGGA |
| RT-PCR miR-2355-3p | RT: GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACATCTCC F: GCCATTGTCCTTGCTGTTT |
R: CAGTGCGTGTCGTGGA |
| RT-PCR miR-369-3p | RT: GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACAAAGAT F: GCCGAATAATACATGGTTG |
R: CAGTGCGTGTCGTGGA |
| RT-PCR miR-9-3p | RT: GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACACTTTC F: GCCGCATAAAGCTAGATAACC |
R: CAGTGCGTGTCGTGGA |
| RT-PCR U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
| RT-PCR IGF2R | CTGCCGCTATGAAATTGAGTGG | CGCCGCTCAGAGAACAAGTT |
| RT-PCR β-actin | TTCCAGCCTTCCTTCCTGGG | TTGCGCTCAGGAGGAGCAAT |
| agomir-NC | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
| agomir-4306 | UGGAGAGAAAGGCAGUA | CUGCCUUUCUCUCCAUU |
| antagomir-NC | CAGUACUUUUGUGUAGUACAA | |
| antagomir-4306 | UACUGCCUUUCUCUCCA | |
| Wt-IGF2R | AATTCTAGGCGATCGCTCGAGACCTTGTGCCCAGGGTTTTG | ATTTTATTGCGGCCAGCGGCCGCCCGCCGGAAGCTGCCCCA |
| mut-IGF2R | TAATTAGAGAGGCCCTATTTATTGACTTTGACAATTACTCA | TAGGGCCTCTCTAATTAAAATGCTAAAATATATCGGGGT |
| IGF2R-pCDNA3.1 | CTAGCGTTTAAACTTAAGCTTATGGGGGCCGCCGCCGGC | TGCTGGATATCTGCAGAATTCTCAGATGTGTAAGAGGTCCTCGTC |
qRT-PCR
Target cells were collected and isolated for total RNA extraction using a TRIzol total RNA isolation kit (Thermo Fisher Scientific, Grand Island, NY, USA). Then, 5 µL of RNA was obtained and diluted in RNA-free ultrapure water at ×20. The density and purity of the RNA were evaluated. The cDNA template was reverse transcribed in a PCR amplifier, and an ABI7500 quantitative PCR instrument was used for qRT-PCR. PCR was performed using β-actin as the internal control and analyzed using the 2−∆∆Ct method. The specific primers used for qRT-PCR are presented in Table 1. The experiment was conducted three times to obtain averages.
Western blotting
A549 and 95D cells were resuspended in RIPA lysis buffer, placed on ice for 15 minutes, and centrifuged at 12,000 rpm for 10 minutes at 4 °C. Total protein samples were collected after discarding the supernatant. Protein concentration was examined using a Bio-Rad protein detection kit (Bio-Rad, Hercules, CA, USA). Protein samples were loaded onto SDS-PAGE gels and then transferred to polyvinylidene fluoride (PVDF) membranes. The membrane was incubated with antibodies against p-PI3K (1:500, ab182651, Abcam, Cambridge, USA), PI3K (1:500, ab278545, Abcam), p-AKT (1:500, ab38449, Abcam), AKT (1:500, ab8805, Abcam) and IGF2R (1:50,000, ab124767, Abcam) at 4 °C overnight. The membrane was then incubated with HRP-conjugated secondary antibody for 1 hour at room temperature. Finally, the protein signals were visualized using ECL substrates (Millipore, USA).
CCK-8 assay
A CCK-8 assay kit (Dojindo, Japan) was used to detect cell viability. Cells were seeded in 96-well plates at a density of 2×103 cells per well. The cells were then added with 20 µL of CCK-8 (10% in culture medium) at 12 and 48 hours, and the cells were incubated for 4 hours at 37 °C. After agitation for 10 min on a shaker, the absorbance at 490 nm was read using a microplate reader.
Colony formation assay
Transfected A549 and 95D cells (1×104/well) were reseeded in 6-well plates and allowed to stand for 12 hours at 37 °C with 5% CO2. The cells were then resuspended in DMEM containing 10% FBS and cultured in 6-well plates (approximately 200 cells per well) at 37 °C for approximately 2 weeks. After incubation, the medium was aspirated, and the cells were fixed with 70% ethanol and subsequently washed with ice-cold PBS. Finally, the cells were stained with 0.1% crystal violet (Beyotime, Shanghai, China), and the number of colonies containing more than 50 cells were counted under a light microscope (Olympus, Tokyo, Japan).
Cell invasion by Transwell assay
A two-chamber Transwell assay determined the invasion of target cells (Corning Incorporation, New York, NY, USA). Target cells were suspended in a 200 µL serum-free RPMI-1640 medium and added to the upper chamber precoated with Matrigel (500 ng/µL; BD Biosciences, Franklin Lakes, NJ, USA). Complete RPMI-1640 medium (600 µL) was added to the lower chamber. After incubation for 48 hours at 37 °C, cells invading the lower chamber were immediately fixed with methanol. Non-invasive cells in the upper chamber were carefully removed using cotton swabs, while invasive cells in the lower chamber were stained with crystal violet and counted under a microscope. Cell invasion (%) was calculated as the average invasive cells in the experimental group/average migrated cells in the control group × 100%.
Subcutaneous tumor model in nude mice
All animal experiments were approved by the Ethics Committee of Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences & Peking Union Medical College (approval number: IMB-20200315D501), in compliance with People’s Republic of China National Standard guidelines for the care and use of animals. A protocol was prepared before the study without registration. Four-week-old male nude (BALB/c) mice (SJA Laboratory Animal Co., Ltd., Shanghai, China) were used in the experiments. A549 cells (5×107 cells/0.1 mL medium/mouse) transfected with agomir-NC or agomir-4306 were injected into the mice subcutaneously to establish the mouse models (17). When the tumors grew to approximately 100 mm3, the mice were randomly divided into four groups (n=6 in each group): agomir-NC, agomir-4306, agomir-NC+ 6 Gy irradiation, and agomir-4306+ 6 Gy irradiation. The mice in the latter two groups received X-ray irradiation at a total dose of 24 Gy (6 Gy/week). On the 28th day, the tumors were subcutaneously harvested following animal sacrifice by cervical dislocation, and examined for tumor volumes and weight.
Hematoxylin & eosin (H&E) staining
The histopathology examination of the tumor tissue from nude mice was implemented using H&E staining. The tumor tissue samples were placed in a 10% formalin solution, dehydrated in an ethanol gradient, embedded in paraffin, and sliced into cross-sections 4 µm thick. After de-paraffinage, the slices were disposed by using H&E staining. The samples were then observed and photographed under a light microscope (Olympus, Tokyo, Japan).
Dual-luciferase reporter assay
Wild-type and mutant-type IGF2R reporter vectors (wt-IGF2R/mut-IGF2R) were constructed based on the psicheck-2 vector (Promega, Madison, WI, USA). The mutant-type vector contained a 7-bp mutation at the predicted miR-4306 binding site. Reporter vectors were cotransduced into cells with agomir-4306/antagomir-4306, and luciferase activity was determined. The specific sequences are listed in Table 1.
Statistical analyses
Data from 3 independent experiments are expressed as the mean ± SD and were processed using SPSS 20.0 statistical software. Differences between groups were compared using a paired two-tailed Student’s t-test or one-way ANOVA, followed by a Tukey’s post-hoc test. Given the high number of evaluated features, correction for multiple testing was performed using the false discovery rate approach. A P value <0.05 was considered significant.
Results
Selecting miRNA(s) that might affect lung adenocarcinoma
To select miRNAs involved in lung adenocarcinoma development, we analyzed differentially expressed miRNAs between normal and lung adenocarcinoma tissues based on GSE135918. As shown in the volcano plot and hierarchical clustering heatmap (Figure 1A,1B), we observed 2,157 human miRNAs, and 412 miRNA expression levels were significantly different, of which 239 were downregulated, and 173 were upregulated. According to the TCGA-LUAD database, 304 miRNAs of the above 412 miRNAs were detected in the Xena dataset. Simultaneously, we used the Welch two-sample t-test to analyze differentially expressed miRNAs between tumor samples (n=450) and control normal tissue samples (n=45). Of these 304 miRNAs, 29 miRNAs were upregulated, and 4 were downregulated (Figure 1C,1D). Next, by a Cox proportional hazard regression model, these 33 differentially expressed miRNAs from the last two steps were analyzed for their correlation with the overall survival rate in patients with lung adenocarcinoma. As shown in Figure 1E, 16 candidates could predict the overall survival of patients with lung adenocarcinoma. We determined the top five miRNAs in terms of the hazard ratio (miR-4529-3p, miR-4306, miR-2355-3p, miR-369-3p, and miR-9-3p) expressed in a normal human lung epithelial cell line and lung adenocarcinoma cell lines. miR-4306 was the most highly expressed miRNA in A549 and 95D cells compared with normal lung epithelial cell (Figure 1F).
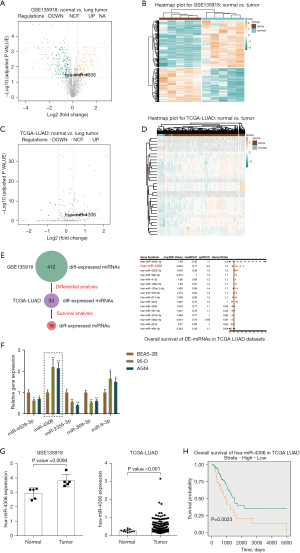
Next, we examined miR-4306 expression in normal and lung adenocarcinoma tissues reported in the GSE135918 and TCGA-LUAD databases. As shown in Figure 1G, the expression level of miR-4306 was increased in cancerous tissue samples. Furthermore, we evaluated the correlation between miR-4306 expression and overall survival of lung adenocarcinoma patients using univariate Cox regression analysis based on the TCGA-LUAD database. As shown in Figure 1H, a higher expression of miR-4306 could predict worse survival in patients with lung adenocarcinoma (P value =0.0023).
In vitro effects of miR-4306 on lung adenocarcinoma cell phenotypes by regulating the PI3K/AKT pathway
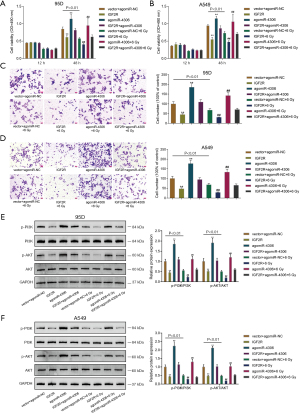
Because higher miR-4306 expression was correlated with a worse prognosis, the specific effects of miR-4306 expression on lung adenocarcinoma cell phenotypes were determined. In 95D and A549 cells, the expression level of miR-4306 was dramatically increased compared with that in BEAS-2B cells (Figure 2A). We exposed 95D and A549 cells to 0, 2, 4, 6, 8, and 10 Gy irradiation; as shown in Figure 2B, the expression of miR-4306 was increased in both cell lines in a dose dependent manner. Then, miR-4306 overexpression or inhibition was achieved in 95D and A549 cells by transducing agomir-4306 or antagomir-4306; miR-4306 overexpression or inhibition was verified by qRT-PCR (Figure 2C). 95D and A549 cells were transduced with agomir-NC/agomir-4306 or antagomir-NC/antagomir-4306 with or without irradiation treatment, and examined for cell phenotypes. As shown in Figure 2D,2E, irradiation alone resulted in significant suppression of cell viability and cell proliferation. Inhibition of miR-4306 decreased, while overexpression of miR-4306 promoted cancer cell viability and cell proliferation. Furthermore, miR-4306 inhibition further enhanced the suppressive effects of irradiation on lung adenocarcinoma cell viability and cell proliferation, while miR-4306 overexpression attenuated the suppressive effects of irradiation on lung adenocarcinoma cell viability and cell proliferation. Consistently, irradiation alone significantly inhibited cancer cell invasion. In addition, inhibition of miR-4306 decreased, while overexpression of miR-4306 promoted cancer cell invasion. Furthermore, miR-4306 inhibition further enhanced the suppressive effects of irradiation on lung adenocarcinoma cell invasion, while miR-4306 overexpression attenuated the suppressive effects of irradiation on lung adenocarcinoma cell invasion (Figure 3A,3B). The effect of miR-4306 on the PI3K/AKT pathway was detected (Figure 3C). Overexpression of miR-4306 promoted, whereas single miR-4306 inhibition markedly decreased, phosphorylated PI3K (p-PI3K) and AKT (p-AKT) protein expression in both 95D and A549 cells. Thus, miR-4306 overexpression attenuated the suppressive effects of irradiation on lung adenocarcinoma cell malignant behaviors through the PI3K/AKT signaling pathway.
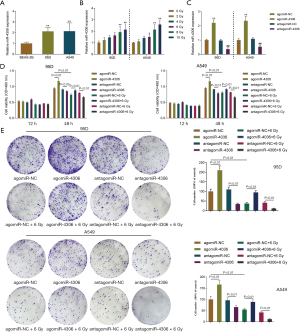
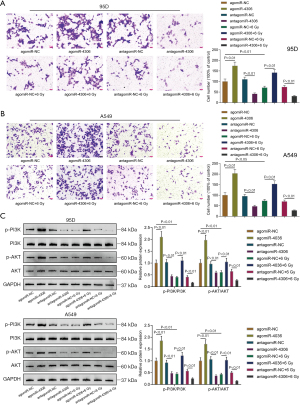
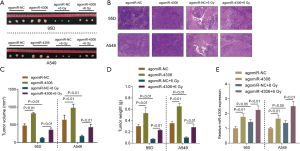
In vivo effects of miR-4306 overexpression on subcutaneously planted tumor growth in nude mice
To further confirm the effects of miR-4306, a subcutaneous tumor model in nude mice was established as described and part of the mouse model received irradiation treatment. Subcutaneous tumors are shown in Figure 4A. H&E staining was then performed to examine the histopathological characteristics of the tumors (Figure 4B). miR-4306 overexpression is correlated with tumor volume and weight (Figure 4C,4D). Irradiation treatment significantly decreased tumor volume and weight, while miR-4306 overexpression attenuated the in vivo effects of irradiation. In tumor tissues, miR-4306 expression was promoted by irradiation, while miR-4306 overexpression increased the stimulative effects of irradiation on miR-4306 (Figure 4E).
miR-4306 directly targets IGF2R
miRNAs exert their biological functions by targeting downstream target mRNAs; therefore, we searched for possible downstream targets of miR-4306. Using miRDIP, miRDB, TargetScan, and mirTarbase, we predicted possible downstream targets of miR-4306, and the prediction results intersected in 10 candidates as follows: LPCAT3, SF1, CCND2, SPATA2, MYBL1, ZC3H11A, CAPZB, IGF2R, PRRT2, and ZNF385A (Figure 5A). Among these 10 candidates, IGF2R attracted our attention for its previously reported tumor suppressive role in lung cancer. IGF2R is frequently down-regulated or mutated in lung cancer [squamous cell carcinoma (18) and adenocarcinoma (19)]. siRNA-induced IGF2R knockdown enhanced the capacity of non-small cell lung cancer cells to proliferate, migrate and invade, while reduced the rate of apoptosis. Moreover, IGF2R knockdown significantly enhanced the cisplatin chemoresistance of non-small cell lung cancer cell lines (18). The IGF2R mutation could lead to the failure of TGF-β-induced suppression of cancer cell proliferation (19). Therefore, we chose IGF2R for further experiments.
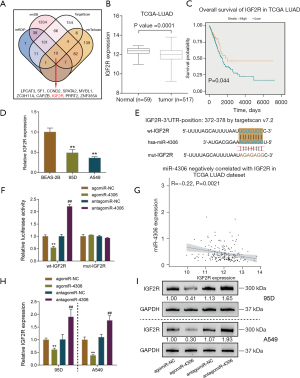
The expression level of IGF2R was dramatically down-regulated in cancerous samples (n=517) compared to non-cancerous samples (n=59), according to the TCGA-LUAD database (Figure 5B). TCGA-LUAD cases were divided into high and low IGF2R groups according to the median expression value of IGF2R. As shown in Figure 5C, higher expression of IGF2R could predict better survival in lung adenocarcinoma patients (Figure 5C). In the 95D and A549 cell lines, the expression level of IGF2R was also dramatically downregulated compared with that in BEAS-2B cells (Figure 5D).
To validate the predicted binding of miR-4306 to IGF2R, we constructed wild-type and mutant IGF2R reporter vectors based on the psiCheck-2 vector. Within the putative miR-4306 binding site of mutant-type reporter vectors, 7 bases were mutated (Figure 5E). Then, we cotransfected cells with agomir-4306 or antagomir-4306. When cotransfected with a wild-type vector, the overexpression of miR-4306 decreased luciferase activity, while the inhibition of miR-4306 promoted luciferase activity. When cotransfected with a mutant-type vector, miR-4306 expression failed to change the luciferase activity (Figure 5F). In tissue samples, miR-4306 and IGF2R expression were negatively correlated, according to the TCGA-LUAD database (Figure 5G). In both 95D and A549 cells, miR-4306 overexpression inhibited IGF2R expression, while miR-4306 inhibition promoted IGF2R expression (Figure 5H,5I).
miR-4306 modulates the response of lung adenocarcinoma cells to irradiation through the IGF2R and PI3K/AKT pathways
After confirming the binding of miR-4306 to IGF2R, the dynamic effects of the miR-4306/IGF2R axis on lung adenocarcinoma cell phenotypes were determined. 95D and A549 cells were divided into 8 groups and transduced accordingly: vector+ agomiR-NC, IGF2R, agomiR-4306, IGF2R+ agomiR-4306, vector+ agomiR-NC+ 6 Gy, IGF2R+ 6 Gy, agomiR-4306+ 6 Gy, and IGF2R+ agomiR-4306+ 6 Gy. Then, cell viability and cell invasion were determined. Without irradiation, IGF2R overexpression inhibited, while miR-4306 overexpression promoted cell viability and invasive capacity. IGF2R overexpression partially attenuated the effects of miR-4306 overexpression on cell viability and invasive capacity (Figure 6A-6D). Under irradiation, the suppressive functions of irradiation in the viability and invasive capacity of cancer cells were enhanced by overexpression of IGF2R, but attenuated by overexpression of miR-4306; the effects of miR-4306 overexpression were also partially attenuated by overexpression of IGF2R (Figure 6A-6D). Then, the effect of miR-4306 along with IGF2R on the expression of the PI3K/AKT pathway related protein was determined (Figure 6E,6F). IGF2R overexpression dramatically suppressed, while miR-4306 overexpression promoted the p-PI3K and p-AKT protein expression in both 95D and A549 cells.
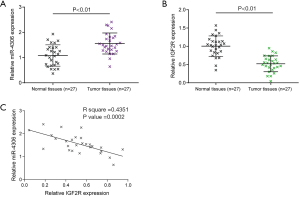
Expression and correlation of miR-4306 and IGF2R in tissue samples
Finally, the expression of miR-4306 and IGF2R in tissues of lung adenocarcinoma (n=27) and the corresponding normal tissues (n=27) was examined. Figure 7A,7B show that miR-4306 expression was upregulated, while IGF2R expression was downregulated in lung adenocarcinoma tissues. In tissue samples, miR-4306 and IGF2R expression were negatively correlated (Figure 7C).
Discussion
This study demonstrated the effect of the miR-4306/IGF2R axis on malignant behaviors of lung adenocarcinoma cells and their response to irradiation. The expression level of miR-4306 was dramatically upregulated in lung adenocarcinoma samples and cells, and could be induced by irradiation in a dose-dependent manner. In lung adenocarcinoma cells, miR-4306 overexpression significantly promoted cell viability and invasive abilities, and attenuated the inhibitory effects of irradiation on cancer cell malignant behaviors. In a subcutaneous tumor model in nude mice, miR-4306 overexpression promoted tumor growth and attenuated the suppressive effects of irradiation on tumor growth. By direct targeting, miR-4306 inhibited the expression of IGF2R. In lung adenocarcinoma cells without irradiation, IGF2R overexpression was inhibited, while IGF2R knockdown promoted cell viability and invasive abilities. The effects of miR-4306 overexpression were partially attenuated by IGF2R overexpression. In lung adenocarcinoma cells with irradiation, suppressive roles of irradiation on the viability and invasive abilities of cancer cells were enhanced through overexpression of IGF2R, but attenuated via IGF2R knockdown; the effects of miR-4306 overexpression were also partially attenuated by overexpression of IGF2R. In tissue samples, miR-4306 and IGF2R were negatively correlated.
Several previous studies have reported miRNA expression patterns within lung adenocarcinoma. Tian et al. (20) analyzed differentially expressed miRNAs in the tumor, adjacent, and normal tissue samples of lung adenocarcinoma. They identified 389 miRNAs, among which miR-21-5p and miR-196a-5p were progressively increased from normal to adjacent to tumor tissue samples, whereas miR-218-5p was progressively decreased in lung adenocarcinoma tissue samples. In particular, miRNAs, including miR-34 (21), miR-320a (22), and miR-198 (23), have been reported to modulate the response of lung cancer cells to radiotherapy. In the present study, by comparing differentially expressed miRNAs between lung adenocarcinoma and normal samples based on the GEO and TCGA-LUAD datasets, we identified miR-4306 as a potential candidate to predict a worse survival probability in patients with lung adenocarcinoma. Previously, miR-4306 expression was increased in multiple cancers, including pancreatic cancer (24) and papillary thyroid cancer (25). In the present study, aberrant upregulation of miR-4306 was observed in lung adenocarcinoma samples. Interestingly, miR-4306 up-regulation in triple-negative breast cancer has been considered a protective factor (26), which could be attributed to regulation via ER-α, HER2, and PR.
As mentioned above, the deregulation of miRNAs in lung adenocarcinoma has been frequently observed. Consistent with the deregulation, miRNAs can modulate cancer cell phenotypes, including proliferation, apoptosis, invasion, migration, and response to chemo- and radiotherapy (8,12). In lung adenocarcinoma, miRNAs, including miR-210 (27), miR-155 (28), miRNA-4315 (29), and miR-1290 (30), have been reported as oncogenic miRNAs promoting cancer cell proliferation, invasion, migration, and resistance to chemotherapy or radiation therapy, and inhibiting apoptosis. Although aberrant up-regulation of miR-4306 has been reported in other cancers (24,25), this is the first definitive demonstration of the oncogenic effects of miR-4306 overexpression in lung adenocarcinoma cells, which promote cancer cell proliferation and invasion, and attenuate cancer cell response to irradiation. More importantly, the role of miR-4306 overexpression in promoting tumor growth and attenuating the suppressive effects of irradiation on tumor growth was validated in vivo.
miRNAs exert their biological functions by regulating gene expression by mRNA decay and/or translational inhibition (5). In this study, we confirm that miR-4306 could directly target IGF2R. As previously mentioned, IGF2R is frequently down-regulated or mutated in lung cancer, including squamous cell carcinoma (18) and adenocarcinoma (19). Low IGF2R expression promoted the ability of non-small cell lung cancer cells to proliferate, migrate, and invade, reduced the rate of apoptosis, and induced the cisplatin chemoresistance of non-small cell lung cancer cells (18). The IGF2R mutation could lead to the failure of TGF-β-induced suppression of cancer cell proliferation (19). In the present study, IGF2R overexpression exerted opposite effects on lung adenocarcinoma cell proliferation, invasion, and response to irradiation to those of miR-4306 overexpression. More importantly, overexpression of IGF2R partially attenuated the oncogenic effects of overexpression of miR-4306 on lung adenocarcinoma cells, suggesting that miR-4306 exerts its functions by targeting IGF2R. In tissue samples, miR-4306 and IGF2R expression was negatively correlated, suggesting the miR-4306/IGF2R axis in lung adenocarcinoma tissues. Impact of these findings on the management of lung adenocarcinoma needs further exploration clinically.
Conclusions
In conclusion, the miR-4306/IGF2R axis could significantly affect lung adenocarcinoma progression and response to radiotherapy, which needs further investigation in vivo and clinically. These findings provide new insight into the underlying mechanisms of lung adenocarcinoma progression and highlight miR-4306 as a potential biomarker for prognosis and radiation sensitivity as well as therapeutic target for lung adenocarcinoma.
Acknowledgments
The authors appreciate the academic support from AME Lung Cancer Collaborative Group and Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences & Peking Union Medical College.
Funding: This study was supported by the National Natural Science Foundation of China (Grant No. 81803060).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/tlcr-21-890
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tlcr-21-890
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tlcr-21-890). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were approved by Research Ethics Committee of the Beijing Hospital, National Center of Gerontology, Institute of Geriatric Medicine, Chinese Academy of Medical Sciences (Approval number: 2018BJYYEC-221-01) and were conducted in accordance with the Helsinki declaration (as revised in 2013). Written informed consent was obtained from all patients. All animal experiments were approved by the Ethics Committee of Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences & Peking Union Medical College (Approval number: IMB-20200315D501), in compliance with People’s Republic of China National Standard guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Butnor KJ. Controversies and challenges in the histologic subtyping of lung adenocarcinoma. Transl Lung Cancer Res 2020;9:839-46. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Scotti V, Bruni A, Francolini G, et al. Stereotactic Ablative Radiotherapy as an Alternative to Lobectomy in Patients With Medically Operable Stage I NSCLC: A Retrospective, Multicenter Analysis. Clin Lung Cancer 2019;20:e53-61. [Crossref] [PubMed]
- Li Z, Yu X, Shen J, et al. MicroRNA dysregulation in uveal melanoma: a new player enters the game. Oncotarget 2015;6:4562-8. [Crossref] [PubMed]
- Gorur A, Balci Fidanci S, Dogruer Unal N, et al. Determination of plasma microRNA for early detection of gastric cancer. Mol Biol Rep 2013;40:2091-6. [Crossref] [PubMed]
- Han D, Li L, Ge X, Li D, et al. MicroRNA expression integrated analysis and identification of novel biomarkers in small cell lung cancer: a meta-analysis. Transl Cancer Res 2020;9:3339-53. [Crossref]
- Niyazi M, Pitea A, Mittelbronn M, et al. A 4-miRNA signature predicts the therapeutic outcome of glioblastoma. Oncotarget 2016;7:45764-75. [Crossref] [PubMed]
- Ma W, Yu J, Qi X, et al. Radiation-induced microRNA-622 causes radioresistance in colorectal cancer cells by down-regulating Rb. Oncotarget 2015;6:15984-94. [Crossref] [PubMed]
- Qu C, Liang Z, Huang J L, et al. MiR-205 determines the radioresistance of human nasopharyngeal carcinoma by directly targeting PTEN. Cell Cycle 2012;11:785-96.
- Wang R, Chen DQ, Huang JY, et al. Acquisition of radioresistance in docetaxel-resistant human lung adenocarcinoma cells is linked with dysregulation of miR-451/c-Myc-survivin/rad-51 signaling. Oncotarget 2014;5:6113-29. [Crossref] [PubMed]
- Summerer I, Niyazi M, Unger K, et al. Changes in circulating microRNAs after radiochemotherapy in head and neck cancer patients. Radiat Oncol 2013;8:296. [Crossref] [PubMed]
- Martin-Kleiner I, Gall Troselj K. Mannose-6-phosphate/insulin-like growth factor 2 receptor (M6P/IGF2R) in carcinogenesis. Cancer Lett 2010;289:11-22. [Crossref] [PubMed]
- Takeda T, Komatsu M, Chiwaki F, et al. Upregulation of IGF2R evades lysosomal dysfunction-induced apoptosis of cervical cancer cells via transport of cathepsins. Cell Death Dis 2019;10:876. [Crossref] [PubMed]
- Zhang Z, Mou Z, Xu C, et al. Autophagy-associated circular RNA hsa_circ_0007813 modulates human bladder cancer progression via hsa-miR-361-3p/IGF2R regulation. Cell Death Dis 2021;12:778. [Crossref] [PubMed]
- Karkare S, Allen KJH, Jiao R, et al. Detection and targeting insulin growth factor receptor type 2 (IGF2R) in osteosarcoma PDX in mouse models and in canine osteosarcoma tumors. Sci Rep 2019;9:11476. [Crossref] [PubMed]
- Han ML, Zhao YF, Tan CH, et al. Cathepsin L upregulation-induced EMT phenotype is associated with the acquisition of cisplatin or paclitaxel resistance in A549 cells. Acta Pharmacol Sin 2016;37:1606-22. [Crossref] [PubMed]
- Tian Z, Yao G, Song H, et al. IGF2R expression is associated with the chemotherapy response and prognosis of patients with advanced NSCLC. Cell Physiol Biochem 2014;34:1578-88. [Crossref] [PubMed]
- Gemma A, Hosoya Y, Uematsu K, et al. Mutation analysis of the gene encoding the human mannose 6-phosphate/insulin-like growth factor 2 receptor (M6P/IGF2R) in human cell lines resistant to growth inhibition by transforming growth factor beta(1) (TGF-beta(1)). Lung Cancer 2000;30:91-8. [Crossref] [PubMed]
- Tian F, Li R, Chen Z, et al. Differentially Expressed miRNAs in Tumor, Adjacent, and Normal Tissues of Lung Adenocarcinoma. Biomed Res Int 2016;2016:1428271. [Crossref] [PubMed]
- Cortez MA, Ivan C, Valdecanas D, et al. PDL1 Regulation by p53 via miR-34. J Natl Cancer Inst 2015;108:djv303. [PubMed]
- Lv Q, Hu JX, Li YJ, et al. MiR-320a effectively suppresses lung adenocarcinoma cell proliferation and metastasis by regulating STAT3 signals. Cancer Biol Ther 2017;18:142-51. [Crossref] [PubMed]
- Liang Y, Wang H, Sun Y, et al. miR-198-induced upregulation of Livin may be associated with the prognosis and contribute to the oncogenesis of lung adenocarcinoma. Oncol Rep 2017;38:2096-104. [Crossref] [PubMed]
- Madhavan B, Yue S, Galli U, et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int J Cancer 2015;136:2616-27. [Crossref] [PubMed]
- Dai D, Tan Y, Guo L, et al. Identification of exosomal miRNA biomarkers for diagnosis of papillary thyroid cancer by small RNA sequencing. Eur J Endocrinol 2020;182:111-21. [Crossref] [PubMed]
- Zhao Z, Li L, Du P, et al. Transcriptional Downregulation of miR-4306 serves as a New Therapeutic Target for Triple Negative Breast Cancer. Theranostics 2019;9:1401-16. [Crossref] [PubMed]
- Xie S, Liu G, Huang J, et al. miR-210 promotes lung adenocarcinoma proliferation, migration, and invasion by targeting lysyl oxidase-like 4. J Cell Physiol 2019;234:14050-7. [Crossref] [PubMed]
- Gu S, Lai Y, Chen H, et al. miR-155 mediates arsenic trioxide resistance by activating Nrf2 and suppressing apoptosis in lung cancer cells. Sci Rep 2017;7:12155. [Crossref] [PubMed]
- Guyon N, Garnier D, Briand J, et al. Anti-PD1 therapy induces lymphocyte-derived exosomal miRNA-4315 release inhibiting Bim-mediated apoptosis of tumor cells. Cell Death Dis 2020;11:1048. [Crossref] [PubMed]
- Wu L, Liu T, Xiao Y, et al. Polygonatum odoratum lectin induces apoptosis and autophagy by regulation of microRNA-1290 and microRNA-15a-3p in human lung adenocarcinoma A549 cells. Int J Biol Macromol 2016;85:217-26. [Crossref] [PubMed]


