
The relationship between different subtypes of KRAS and PD-L1 & tumor mutation burden (TMB) based on next-generation sequencing (NGS) detection in Chinese lung cancer patients
Introduction
Lung cancer is a leading cause of cancer-related deaths worldwide (1). KRAS, the most common oncogene, has been found in 26.1% of lung adenocarcinomas (LADCs) and 6.4% of squamous cell carcinomas (SQCCs) in Western countries and in 11.2% and 1.8% of lung cancer cases in Asia, respectively (2,3). Tumors with KRAS mutation are some of the most invasive and refractory types of tumors. KRAS mutations have a higher prevalence in Western countries and in smokers (4).
Targeted drugs have been developed for non-small cell lung cancer (NSCLC) to inhibit tumor proliferation and invasion. The discovery of targeted therapies for patients with epidermal growth factor receptor (EGFR) mutation and anaplastic lymphoma kinase (ALK) or c-ros oncogene 1 (ROS1) rearrangements has been a breakthrough in the treatment history of NSCLC (5-9). However, for many years, effective KRAS-targeted therapies have been limited with only recent approvals for the KRAS G12C isoform in lung cancer.
The RAS gene family encodes enzymes to hydrolyze GTPase, linking upstream cell surface receptors such as EGFR and fibroblast growth factor receptor (FGFR) to downstream survival and proliferation pathways, including Raf/mitogen-activated protein kinase (MEK)/extracellular signal-regulated kinase (ERK), phosphatidylinositol-3 kinase (PI3K)/protein kinase B (AKT)/mammalian target of rapamycin (mTOR), and Ral guanine nucleotide dissociation stimulator (RalGDS)/RA (4). As the most common oncogene, KRAS, HRAS, and NRAS mutations occur in 30% of cancer cases. KRAS is the most common mutation subtype, present in 86% of RAS mutations in cancer, followed by NRAS (11%), and HRAS (3%) (8). RAS has the highest modification rate in lung, pancreatic, and colorectal cancer, and KRAS is the most common in lung, pancreas, and colon cancer (8). KRAS mutations usually occur at codons 12, 13, and 61, and the most common codon mutations include G12C (GGT to TGT), G12V (GGT to GTT), and G12D (10). The subtypes of KRAS have been shown to drive different biological characteristics (11). Patients with p.G12V or p.G12C have been reported to have longer survival than those with other types of mutations (12-14). Patients who have never smoked are more common in G12D subtypes. The transition mutations (G→T or G→C) are more common in smokers, and transition mutations (G→A) are more common in never-smokers.
KRAS mutation is classically defined as a negative prognostic factor. KRAS-mutant lung cancer has poorer survival and disease-free survival compared to KRAS wild-type tumors in both early stage and advanced metastatic lung cancer patients, indicating the urgent need for novel treatment strategies for KRAS-driven NSCLC (15). The KRAS G12C inhibitor sotorasib (AMG510) was recently approved by the US Food and Drug Administration (FDA) (16-18). The current standard options for patients with KRAS mutations include chemotherapy and/or checkpoint immunotherapy. Immunotherapy has advanced tumor treatment strategies, and patients with KRAS mutations may in some cases have increased immune response. The oncogene-specific differences are demonstrated in the expression of TMB and PD-L. NSCLC with BRAF mutations demonstrated superior benefit of ICB, which may be attributed to higher TMB and higher PD-L1 expression (19). A systematic meta-analysis demonstrated that patients with KRAS mutations show clinical benefits from the anti-PD-1/PD-L1 immunotherapy (20). Studies have shown that KRAS mutations are associated with the inflammatory tumor microenvironment and tumor immunogenicity, leading to better patient responses to PD-1 inhibitors. Subgroup analyses of clinical trial have indicated that the KRAS-mutant patients are more sensitive to PD-1/PD-L1 inhibitors than the wild-type (21-23). However, two recent studies have provided further insights into the predictive potential of KRAS mutations, concluding that KRAS Mutation status did not differ significantly in objective response rate (ORR), progression-free survival (PFS), or OS. The different subtypes of KRAS mutations may be associated with distinct therapeutic effects, and some may not benefit from immunotherapy (24,25). About half of KRAS-mutant NSCLCs have been found to harbor concomitant mutations, including TP53, STK11, and CDKN2A/B (26,27). One study demonstrated the potential predictive value of TP53 and KRAS mutation for response to programmed cell death 1 (PD-1) blockade immunotherapy in lung adenocarcinoma (28). Objective response rates to PD-1 blockade differed significantly among STK11/LKB1 & KRAS (7.4%), TP53 & KRAS (35.7%), and KRAS-only (28.6%) (29). It has also been reported that KRAS G12D mutation predicts lower tumor mutation burden (TMB) and indicates immune suppression in LADC.
Therefore, we aimed to explore the expression level of TMB and programmed death-ligand 1 (PD-L1) in patients with KRAS mutant subtypes, providing more evidence for the response of KRAS mutant subtypes to immunotherapy. We conducted analysis of 2880 KRAS-mutant NSCLC patients who had PD-L1 testing performed and next generation sequencing performed for TMB testing.
We present the following article in accordance with the MDAR reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-88/rc).
Methods
Data collection and preparation
A total of 2,880 NSCLC patients in East China were included in the study. The data were provided by Berry Oncology (Fujian, China), Geneplus BioTech (Beijing, China), Nanjing Geneseeq Technology Inc (Nanjing, China), and Burning Rock Biotech (Guangzhou, China). TMB expression was defined by the number of somatic mutations (besides intron mutation and synonymous mutation) per genome area for target sequencing (38 Mb). Somatic mutations were defined as nonsynonymous mutations, non-silent mutations, deletion mutations, insertion mutations, and frameshift mutations. The PD-L1 clone 22C3 pharmDx kit was used to measure the expression level of PD-L1.
The study was approved by the institutional review board at Shanghai Jiao Tong University, Shanghai Chest Hospital (No. IS21126). It was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Individual consent for this retrospective analysis was waived.
Statistical analyses
Z-scores were calculated by (x−µ)/σ to convert 2 or more sets of data into unitless scores. SPSS 20.0 (SPSS, Chicago, IL, USA) software was used for statistical analyses. All scatter plots and boxplot maps were drawn using GraphPad Prism 8. Mann-Whitney U test was used for statistical analysis. P value <0.05 was considered statistically significant.
Results
Clinical characteristics of Chinese KRAS-mutant NSCLC patients
To investigate TMB in KRAS-mutant NSCLC, we interrogated detailed somatic mutation data of NSCLC in tissues (2,880 cases) and blood (758 cases). Z-scores were used to unify all data. The prevalence of main mutations in 2880 patients with NSCLC is summarized in Figure 1A. Of the 2880 NSCLC patients harboring KRAS mutations, KRAS p.A146X was detected in 37 (1.2%) patients. The most frequent mutations found in KRAS-MUT patients were p.G12C (n=904, 31.4%) and p.G12D (n=523, 18.2%). The proportions of the other 2 major p.G12 subtypes were G12V, 15.5% (n=447) and G12A, 7.3% (n=210). The proportion of G13C and G13D was 3% (n=87) and 3.3% (n=94), respectively. Q61X accounted for 5.5% of KRAS mutations, and Q61H, Q61K, Q61L, and Q61R accounted for 4.2%, 0.3%, 0.8%, and 0.2%, respectively (Figure 1A).
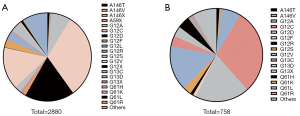
We screened blood samples from 758 NSCLC patients with KRAS mutation. A146X was detected in 13 (1.8%) patients. The most frequent mutations found in KRAS-MUT patients were p.G12C (n=227, 29.9%) and p.G12D (n=144,19%). The proportions of the other 2 major p.G12 subtypes were G12V, 15.7% (n=119) and G12A, 6.7% (n=51). The proportion of G13C and G13D was 3.7% (n=28) and 3.2% (n=24), respectively. Q61X accounted for 6.4% of KRAS mutations, and Q61H, Q61K, Q61L, and Q61R accounted for 4.4%, 0.4%, 1.1%, and 0.5%, respectively (Figure 1B).
TMB in tissue across KRAS genotypes
TMB expression was analyzed in the tissues of 2880 KRAS-MUT patients, including A146X (Z-score of TMB =0.04), A59X (Z-score of TMB= 0.37), G12X (Z-score of TMB =−0.02), G13X (Z-score of TMB= 0.35), and Q61X (Z-score of TMB =−0.28). Z-scores were calculated by (x−µ)/σ to convert 2 or more sets of data into unitless scores. We found that TMB expression in KRAS p.Q61X mutation was at a lower level (Z-score of TMB =−0.28). Meanwhile, KRAS p.A59X (Z-score of TMB =0.37) and KRAS p.G13X (Z-score of TMB =0.35) showed higher TMB expression (Figure 2A).
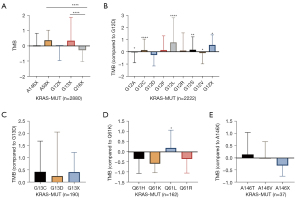
Among the 2,880 patients with KRAS mutation, p.G12X patients accounted for 77.2% (2,222). TMB expression in p.G12L was the highest (Z-score of TMB =0.77), followed by p.G12F (Z-score of TMB =0.14) and p.G12C (Z-score of TMB =0.14). TMB expression in p.G12D was the lowest of p.G12X type (Z-score of TMB =−0.26, Figure 2B). There was no significant difference in TMB expression between p.G13C and p.G13D in patients with p.G13 mutation (n=190) (Figure 2C). Among the patients with p.Q61X mutation (n=162), TMB expression in p.Q61K was the lowest (Z-score of TMB =−0.58) and TMB expression in p.Q61L (Z-score of TMB =0.20) was the highest (p.Q61L vs. p.Q61K, P=0.01) (Figure 2D). There was also no significant difference between p.A146T and p.A146V in patients with A146 mutation (n=37) (Figure 2E).
KRAS Q61X mutation showed decreased TMB in blood
TMB expression was also analyzed in blood samples of 758 KRAS-MUT patients, including A146X (Z-score of TMB =−0.15), G12X (Z-score of TMB =−0.06), G13X (Z-score of TMB =0.50), and Q61X (Z-score of TMB =−2.6). The expression level of TMB in G13X was the highest, and TMB expression in Q61X was the lowest. Patients with G13X mutation might have a better response to immunotherapy, while patients with Q61X mutation may have poorer response to immunotherapy (Figure 3A).
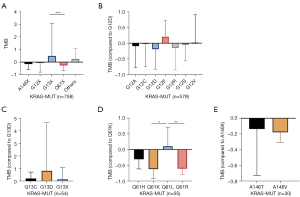
TMB expression in p.G12D (Z-score of TMB =−1.9) was the lowest of the p.G12 mutation type. There was no significant difference among genotype in blood samples (Figure 3B). There was no significant difference in TMB expression between G13C and G13D in patients with G13 mutation (n=54) (Figure 3C). Among the patients with Q61 mutation (n=55), TMB expression in Q61K (Z-score of TMB =−0.62) was the lowest, and TMB expression in Q61L (Z-score of TMB =0.11) was the highest (Q61L vs. Q61K P=0.01) (Figure 3D). There was also no significant difference between A146T and A146V in patients with A146 mutation (n=30) (Figure 3E). In conclusion, the blood analysis results provide support for the conclusions of the tissue analysis.
KRAS Q61X mutation showed decreased PD-L1 in tissue
The expression of PD-L1 in NSCLC patients with KRAS mutation was also screened (Figure S1, n=412), including A146X (TPS =16.33%), G12X (TPS =27.25%), G13X (TPS =35.21%), and Q61X (TPS =12.82%) (Figure 4A). The expression of PD-L1 in the Q61X mutation was at a lower level (TPS =12.82%) (Figure 4B,4C).
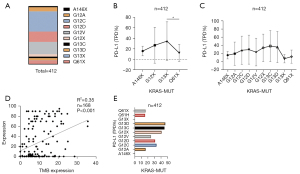
A positive correlation between TMB and PD-L1 was confirmed (Figure 4D). The proportion of patients with PD-L1 expression greater than 50% in each KRAS mutation subtype was further analyzed. The proportion of patients with high PD-L1 [tumor proportion score (TPS) ≥50%] expression in G13X and Q61X was 46.43% (n=13/28) and 16% (n=4/25), respectively (Figure 4E). In conclusion, the proportion of patients with high PD-L1 expression in G13X-MUT was significantly higher than that of Q61X-MUT patients.
The composition ratio of G13X in KRAS/TP53 co-mutation is significantly higher than Q61X in tissue
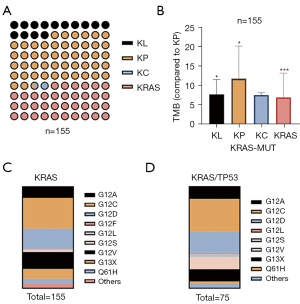
The clinical data from Burning Rock Biotech (Guangzhou, China) were analyzed (n=155). The patients were divided into 4 groups: KRAS/TP53, KRAS/STK11, KRAS/CDKN2A, and KRAS mutation (without TP53, STK11, and CDKN2A), respectively. The most common mutation subtype was KRAS/TP53 (n=75, 48.39%), followed by KRAS (n=56, 36.13%), KRAS/STK11 (n=22, 14.19%), and KRAS/CDKN2A (n=2, 1.29%) (Figure 5A). Analysis of TMB expression in the 4 groups showed that it was highest in KRAS/TP53 (Figure 5B). We further analyzed the proportion of subtypes in the KRAS and KRAS/TP53 groups in tissue samples of 155 NSCLC patients. The proportion of G13X in the KRAS and KRAS/TP53 groups was 9.6% and 12%, respectively (Figure 5C,5D). The proportion of Q61H in the KRAS and KRAS/TP53 was 4.5% and 2.6%, respectively (Figure 5C,5D). In conclusion, the composition ratio of G13X in KRAS/TP53 co-mutation was significantly higher than the ratio of Q61H in KRAS mutation patients.
The composition ratio of G13X in KRAS/TP53 co-mutation is significantly higher than Q61X in blood
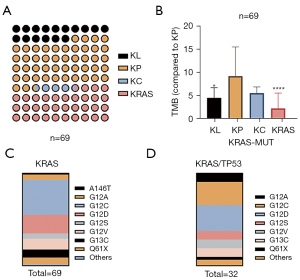
The clinical data of 69 patients with KRAS mutation were analyzed. The most common mutation subtype was KRAS/TP53 (n=32, 46.38%), followed by KRAS (n=23, 33.33%), KRAS/STK11 (n=11, 15.94%), and KRAS/CDKN2A (n=3, 4.35%) (Figure 6A). TMB expression in KRAS/TP53 was the highest (specify) (Figure 6B). We further analyzed the proportion of subtypes in the KRAS and KRAS/TP53 groups in blood samples of 69 NSCLC patients. The proportion of G13X in the KRAS and KRAS/TP53 groups was 8.7% and 10%, respectively (Figure 6C,6D). The proportion of Q61H in the KRAS and KRAS/TP53 groups was 7.4% and 3.1%, respectively (Figure 6C,6D).
Discussion
KRAS mutation occurs in 20–40% of LADCs (30). RAS signaling is associated with various immune-modulating effects, including the regulation of CD8+ lymphocyte infiltration, PD-L1 expression, and myeloid-derived suppressor cell density in the tumor microenvironment (TME) (31-34). Smoking and KRAS mutation were positively associated with the expression of PD-L1 on immune cells. Multivariate analysis demonstrated that KRAS mutation and smoking were independent predictors for positive expression of PD-L1 on immune cells (35,36). Patients with KRAS mutation and extensive smoking history were more likely to have PD-L1 expression on both tumor cells and immune cells (37). Of patients with KRAS mutation, G12D has a higher proportion of never-smokers, and smoking is associated with high PD-L1 expression. The patients with KRAS G12D mutation were found to have markedly decreased TMB and PD-L1 expression, which was consistent with the conclusions of previous research.
Different KRAS mutation subtypes have been confirmed to have different biological characteristics. Three common G12 mutations are reported to be associated with poor outcomes, including G12C, G12V, and G12R (38,39). In particular, mutations in G12C and G12V are associated with worse survival compared with other KRAS mutation subtypes (40). There was no statistically significant difference in overall survival (OS) between the 3 main subtypes (G12C vs. G12D vs. G12V, P=0.81) and codons (12 vs. 13 vs. 61, P=0.36) of KRAS mutations. Codon 13 had a lower estimated 2-year OS rate of 38.6% (95% CI: 21.0–55.9) and codon 61 had a relatively higher 2-year OS rate of 65.0% (95% CI: 42.5–80.5) (14).
On May 28, 2021, the FDA granted accelerated approval for sotorasib (Lumakras™, Amgen, Inc.), a RAS GTPase-family inhibitor, for patients with KRAS G12C-mutated locally advanced or metastatic NSCLC who have received at least 1 prior systemic therapy (18). However, the majority of KRAS mutations remain not directly targetable (41). Therefore, we aimed to explore the immunological properties of different KRAS subtypes. We demonstrated that patients with KRAS Q61 mutation and patients with KRAS G13 mutation had markedly decreased TMB and PD-L1 expression.
Mutations in KRAS, mesenchymal-epithelial transition (MET), and TP53 were significantly correlated with high PD-L1 expression (each P<0.001, q<0.001), while STK11 and EGFR mutations correlated with low PD-L1 expression (P<0.001, respectively) (42). KRAS mutation is associated with an increase in the ratio of PD-L1 and CD8+ tumor-infiltrating lymphocytes (TILs), increased TMB, and immunogenicity (20). KRAS mutations were divided into 3 subgroups according to the co-mutation types in KRAS-mutated lung cancer: KRAS/TP53 (KP), KRAS/STK11 (KL), and KRAS/CDKN2A (KC) (26,28,43,44). KL tumors demonstrated lower levels of immune checkpoint effector molecules, such as PD-L1. KP tumors expressed higher levels of immune markers, somatic mutations, inflammatory markers, and improved recurrence-free survival (45). However, whether the mutant subtypes of KRAS could predict clinical response to immune checkpoint inhibitions (ICIs) remains unclear. Consistent with previous studies, our study found that TP53/KRAS co-mutation can significantly increase TMB. Meanwhile, we found that the expression ratio of G13 in the KRAS/TP53 group was higher than that in the KRAS group, although this requires further validation in larger datasets.
Several studies and data from major trials have demonstrated the relationship between high TMB and response to ICIs in populations (46). TMB has also been shown to be correlated with better progression-free survival (PFS) of ICIs in NSCLC. In patients treated with PD-1/PD-L1 inhibitors (n=1,290, 31.7%), TMB of 20 or more was associated with superior OS [16.8 months (95% CI: 11.6–24.9) vs. 8.5 months (95% CI: 7.6–9.7), P<0.001] and longer time receiving therapy [7.8 months (95% CI: 5.5–11.1) vs. 3.3 months (95% CI: 2.8–3.7), P<0.001] compared to TMB less than 20 (47). PD-L1 has also been approved by the FDA as a diagnostic marker for checkpoint inhibitors (22,48). However, not all patients with high expression of PD-L1 respond to therapy or demonstrate a durable clinical benefit. KEYNOTE-024 demonstrated better OS for pembrolizumab vs. chemotherapy in patients with high PD-L1 expression (PD-L1 expression ≥50%) (49,50). Therefore, we further explored the proportion of PD-L1 expression in each subtype of KRAS mutation (PD-L1 ≥50%). The proportion of PD-L1 (TPS ≥50%) was 46% and 16% in G13X and Q61X, respectively. This also suggested that in patients with KRAS mutations, the G13X group could achieve better survival through immunotherapy, while the Q61X group gained poorer survival benefit.
The comprehensive evaluation of the expression level of both TMB and PD-L1 may provide greater guidance value for patient response to immunotherapy. We confirmed that G13X subtype expressed both higher levels of TMB and PD-L, while Q61X expressed lower levels of both TMB and PD-L1. Subtype alterations in KRAS-mutant NSCLC may be predictors of anti-PD-1/PD-L1 treatment sensitivity, which could also provide more individualized treatment for patients with KRAS-mutant lung cancer.
Conclusions
Our research confirmed the diversity of genetic backgrounds in different KRAS subtypes. We have demonstrated the low level of TMB and expression of PD-L1 in KRAS Q61X-mutant lung cancer and the high level of TMB and expression of PD-L1 in KRAS G13X-mutant lung cancer. Our conclusions were consistently verified in tissue and blood samples, and supports the feasibility of NGS verification in blood. The subtype of KRAS mutation could be a potential therapeutic biomarker in KRAS-mutant lung cancer, providing better individualized treatment strategies for lung cancer patients.
Acknowledgments
The authors appreciate the academic support from the AME Lung Cancer Collaborative Group. We would like to thank all the colleagues in our research team.
Funding: This study was funded by National Natural Science Foundation of China (No. 82072564), Project of Shanghai Natural Science Foundation (No. 20ZR1452000), Shanghai youth talent support program, Shanghai Chest Hospital Project of Collaborative Innovative Grant (No. YJT20191015), Lian Yun Gang Shi Hui Lan Public Foundation (No. HL-HS2020-65), Guangdong Association of Clinical Trials (GACT)/Chinese Thoracic Oncology Group (CTONG) and Guangdong Provincial Key Lab of Translational Medicine in Lung Cancer (No. 2017B030314120), National Multi-disciplinary Treatment Project for Major Diseases (No. 2020NMDTP).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-88/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-88/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-88/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the institutional review board at Shanghai Jiao Tong University, Shanghai Chest Hospital (No. IS21126). It was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Grommes C, DeAngelis LM. Primary CNS Lymphoma. J Clin Oncol 2017;35:2410-8. [Crossref] [PubMed]
- Jordan EJ, Kim HR, Arcila ME, et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer Discov 2017;7:596-609. [Crossref] [PubMed]
- Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer 2003;3:11-22. [Crossref] [PubMed]
- Heymach JV, Nilsson M, Blumenschein G, et al. Epidermal growth factor receptor inhibitors in development for the treatment of non-small cell lung cancer. Clin Cancer Res 2006;12:4441s-5s. [Crossref] [PubMed]
- Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385-94. [Crossref] [PubMed]
- Drilon A, Wang L, Hasanovic A, et al. Response to Cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov 2013;3:630-5. [Crossref] [PubMed]
- Cox AD, Fesik SW, Kimmelman AC, et al. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov 2014;13:828-51. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Wood K, Hensing T, Malik R, et al. Prognostic and Predictive Value in KRAS in Non-Small-Cell Lung Cancer: A Review. JAMA Oncol 2016;2:805-12. [Crossref] [PubMed]
- Tape CJ, Ling S, Dimitriadi M, et al. Oncogenic KRAS Regulates Tumor Cell Signaling via Stromal Reciprocation. Cell 2016;165:910-20. [Crossref] [PubMed]
- Garrido P, Olmedo ME, Gómez A, et al. Treating KRAS-mutant NSCLC: latest evidence and clinical consequences. Ther Adv Med Oncol 2017;9:589-97. [Crossref] [PubMed]
- Dogan S, Shen R, Ang DC, et al. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res 2012;18:6169-77. [Crossref] [PubMed]
- El Osta B, Behera M, Kim S, et al. Characteristics and Outcomes of Patients With Metastatic KRAS-Mutant Lung Adenocarcinomas: The Lung Cancer Mutation Consortium Experience. J Thorac Oncol 2019;14:876-89. [Crossref] [PubMed]
- Reck M, Carbone DP, Garassino M, et al. Targeting KRAS in non-small-cell lung cancer: recent progress and new approaches. Ann Oncol 2021;32:1101-10. [Crossref] [PubMed]
- Govindan R, Fakih M, Price T, et al. Phase I study of AMG 510, a novel molecule targeting KRAS G12C mutant solid tumours. Ann Oncol 2019;30:v163-4. [Crossref]
- Canon J, Rex K, Saiki AY, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019;575:217-23. [Crossref] [PubMed]
- Palma G, Khurshid F, Lu K, et al. Selective KRAS G12C inhibitors in non-small cell lung cancer: chemistry, concurrent pathway alterations, and clinical outcomes. NPJ Precis Oncol 2021;5:98. [Crossref] [PubMed]
- Negrao MV, Skoulidis F, Montesion M, et al. Oncogene-specific differences in tumor mutational burden, PD-L1 expression, and outcomes from immunotherapy in non-small cell lung cancer. J Immunother Cancer 2021;9:e002891. [Crossref] [PubMed]
- Liu C, Zheng S, Jin R, et al. The superior efficacy of anti-PD-1/PD-L1 immunotherapy in KRAS-mutant non-small cell lung cancer that correlates with an inflammatory phenotype and increased immunogenicity. Cancer Lett 2020;470:95-105. [Crossref] [PubMed]
- Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:1627-39. [Crossref] [PubMed]
- Fehrenbacher L, Spira A, Ballinger M, et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 2016;387:1837-46. [Crossref] [PubMed]
- Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 2017;389:255-65. [Crossref] [PubMed]
- Passiglia F, Cappuzzo F, Alabiso O, et al. Efficacy of nivolumab in pre-treated non-small-cell lung cancer patients harbouring KRAS mutations. Br J Cancer 2019;120:57-62. [Crossref] [PubMed]
- Jeanson A, Tomasini P, Souquet-Bressand M, et al. Efficacy of Immune Checkpoint Inhibitors in KRAS-Mutant Non-Small Cell Lung Cancer (NSCLC). J Thorac Oncol 2019;14:1095-101. [Crossref] [PubMed]
- Skoulidis F, Byers LA, Diao L, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov 2015;5:860-77. [Crossref] [PubMed]
- Huang L, Guo Z, Wang F, et al. KRAS mutation: from undruggable to druggable in cancer. Signal Transduct Target Ther 2021;6:386. [Crossref] [PubMed]
- Dong ZY, Zhong WZ, Zhang XC, et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin Cancer Res 2017;23:3012-24. [Crossref] [PubMed]
- Skoulidis F, Goldberg ME, Greenawalt DM, et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov 2018;8:822-35. [Crossref] [PubMed]
- Ricciuti B, Leonardi GC, Metro G, et al. Targeting the KRAS variant for treatment of non-small cell lung cancer: potential therapeutic applications. Expert Rev Respir Med 2016;10:53-68. [Crossref] [PubMed]
- Adderley H, Blackhall FH, Lindsay CR. KRAS-mutant non-small cell lung cancer: Converging small molecules and immune checkpoint inhibition. EBioMedicine 2019;41:711-6. [Crossref] [PubMed]
- Lee CK, Man J, Lord S, et al. Clinical and Molecular Characteristics Associated With Survival Among Patients Treated With Checkpoint Inhibitors for Advanced Non-Small Cell Lung Carcinoma: A Systematic Review and Meta-analysis. JAMA Oncol 2018;4:210-6. [Crossref] [PubMed]
- Dias Carvalho P, Guimarães CF, Cardoso AP, et al. KRAS Oncogenic Signaling Extends beyond Cancer Cells to Orchestrate the Microenvironment. Cancer Res 2018;78:7-14. [Crossref] [PubMed]
- Uprety D, Adjei AA. KRAS: From undruggable to a druggable Cancer Target. Cancer Treat Rev 2020;89:102070. [Crossref] [PubMed]
- Tseng JS, Yang TY, Wu CY, et al. Characteristics and Predictive Value of PD-L1 Status in Real-World Non-Small Cell Lung Cancer Patients. J Immunother 2018;41:292-9. [Crossref] [PubMed]
- Chen YB, Mu CY, Huang JA. Clinical significance of programmed death-1 ligand-1 expression in patients with non-small cell lung cancer: a 5-year-follow-up study. Tumori 2012;98:751-5. [Crossref] [PubMed]
- Song P, Guo L, Li W, et al. Clinicopathologic Correlation With Expression of PD-L1 on Both Tumor Cells and Tumor-infiltrating Immune Cells in Patients With Non-Small Cell Lung Cancer. J Immunother 2019;42:23-8. [Crossref] [PubMed]
- Yu HA, Sima CS, Shen R, et al. Prognostic impact of KRAS mutation subtypes in 677 patients with metastatic lung adenocarcinomas. J Thorac Oncol 2015;10:431-7. [Crossref] [PubMed]
- Uras IZ, Moll HP, Casanova E. Targeting KRAS Mutant Non-Small-Cell Lung Cancer: Past, Present and Future. Int J Mol Sci 2020;21:4325. [Crossref] [PubMed]
- Ihle NT, Byers LA, Kim ES, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst 2012;104:228-39. [Crossref] [PubMed]
- Drosten M, Barbacid M. Targeting the MAPK Pathway in KRAS-Driven Tumors. Cancer Cell 2020;37:543-50. [Crossref] [PubMed]
- Schoenfeld AJ, Rizvi H, Bandlamudi C, et al. Clinical and molecular correlates of PD-L1 expression in patients with lung adenocarcinomas. Ann Oncol 2020;31:599-608. [Crossref] [PubMed]
- La Fleur L, Falk-Sörqvist E, Smeds P, et al. Mutation patterns in a population-based non-small cell lung cancer cohort and prognostic impact of concomitant mutations in KRAS and TP53 or STK11. Lung Cancer 2019;130:50-8. [Crossref] [PubMed]
- Schabath MB, Welsh EA, Fulp WJ, et al. Differential association of STK11 and TP53 with KRAS mutation-associated gene expression, proliferation and immune surveillance in lung adenocarcinoma. Oncogene 2016;35:3209-16. [Crossref] [PubMed]
- Galan-Cobo A, Sitthideatphaiboon P, Qu X, et al. LKB1 and KEAP1/NRF2 Pathways Cooperatively Promote Metabolic Reprogramming with Enhanced Glutamine Dependence in KRAS-Mutant Lung Adenocarcinoma. Cancer Res 2019;79:3251-67. [Crossref] [PubMed]
- Chan TA, Yarchoan M, Jaffee E, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol 2019;30:44-56. [Crossref] [PubMed]
- Singal G, Miller PG, Agarwala V, et al. Association of Patient Characteristics and Tumor Genomics With Clinical Outcomes Among Patients With Non-Small Cell Lung Cancer Using a Clinicogenomic Database. JAMA 2019;321:1391-9. [Crossref] [PubMed]
- Wu SP, Liao RQ, Tu HY, et al. Stromal PD-L1-Positive Regulatory T cells and PD-1-Positive CD8-Positive T cells Define the Response of Different Subsets of Non-Small Cell Lung Cancer to PD-1/PD-L1 Blockade Immunotherapy. J Thorac Oncol 2018;13:521-32. [Crossref] [PubMed]
- Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2016;375:1823-33. [Crossref] [PubMed]
- Mok TSK, Wu YL, Kudaba I, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet 2019;393:1819-30. [Crossref] [PubMed]


