Prediction of future imagery of lung nodule as growth modeling with follow-up computed tomography scans using deep learning: a retrospective cohort study
Introduction
Multiple national clinical trials have supported that lung cancer screening (LCS) by low-dose computed tomography (LDCT) reduces mortality in high-risk populations with a smoking history and family history of cancer (1). The at-risk population eligible for LCS was estimated at over 8 million in the US (2), and would be several times larger in China. The large-scale screening program imposes a heavy burden on screening personnel to interpret the massive volume of imaging data alongside other clinical data (3,4). It is not only time-consuming to detect lung nodules, the early radiological sign of lung cancer, it is also difficult to determine the malignancy potential with reliable accuracy (5).
In clinical practice, follow-up CT scans are recommended by lung nodule management guidelines (6) and routinely prescribed by radiologist to monitor the growth patterns of lung nodules for which the risk of malignancy cannot be clearly determined on baseline scans. Volume doubling time (VDT) (5) and consolidation-to-tumor ratio (CTR) (7) are 2 imaging-based metrics used in screening settings to characterize the growth patterns and the proportion of solid components of lung nodules, respectively. According to the British Thoracic Society (BTS) guidelines, the VDT of newly found solid nodules of intermediate size (4–8 mm) can be computed to categorize the lung nodule as stable, slow growth (VDT >600 days), or fast growth (VDT <600 days) in the follow-up examination (8). With relatively indolent growth patterns, sub-solid nodules generally have longer VDT, require longer follow-up periods, and are more likely to be benign or preinvasive if VDT >1,500 (9). Based on the growth rate indicated by the VDT, conventional clinical management include discharging the patient for stable nodule, ongoing surveillance for slowly growing nodule, and biopsy or resection for fast growing nodule. Besides VDT, the increase of consolidation component in sub-solid lung nodules is also considered a risk factor for malignancy. This notion has not only been mentioned in the major guidelines of lung nodule management but is also supported by previous studies. The guidelines recommend further investigation for part-solid nodules with increasing solid component found in follow-up scans (6). A retrospective study on patients with stage IA lung cancer found that higher CTR was associated with lymphatic invasion and shorter recurrence-free survival (10).
To alleviate the heavy clinical workload in the LCS program, efforts have been taken to develop machine learning algorithms to automatically detect lung nodules (11) and predict their probability of malignancy (12) on CT scans at scale. Thanks to the breakthrough in computer vision made possible by deep learning, these algorithms have achieved a precision level matching a senior radiologist but with far more efficiency (13). While most of these algorithms were built on data collected at a single time point (14-16), recent studies started to provide evidence on the additive values of using sequential imaging data in the analysis of lung nodules compared to data at single time point (17,18). For instance, the importance of growth properties for nodule characterization prompted the 2018 ISBI challenge on nodule malignancy prediction using sequential CT scans, where the best participants reported higher AUC than the contestants in LUNGx challenge that used CT scans on a single time point as inputs (17). Nonetheless, no consensus has been reached regarding how to incorporate the malignancy scores outputted by these risk prediction models into the clinical workflow of lung nodule management. Only the Brock risk model is taken into consideration for managing part-solid nodules by the guidelines of the BTS (8). As far as the authors know, no advanced model based on deep learning with higher accuracy has been cited in guidelines of any society to date. It is still subject to screening personnel as to how to utilize these risk predictions.
Growth pattern is an intermediate attribute of lung nodule that clinicians make sense of and routinely use as one of the deciding factors for the nodule malignancy. Empirically, nodule growth pattern as indicated by change in volume has been reported a strong predictor for nodule malignancy (19). The nodule growth estimate thus may be of greater clinical value for clinicians compared to the malignancy score. Nevertheless, the modeling of nodule growth was rarely studied, and the large volume of follow-up CT scans were still under exploited. In the literature, we only found few related studies on this topic. Shi et al constructed binary classifier of nodule growth using quantitative imaging features at baseline CT scans (20), which did not provide any visual clue for clinicians to inspect. Sheng et al reconstructed the image patches of lung nodule in a future scan point via predicted registration field based on the visual difference between previous two scan points (21). Although the reconstructed image was encouraging, their goal was to predict the nodule malignancy based on the reconstructed image patch with another neural network. While the reconstructed image patch of nodule was informative to clinicians, the growth properties such as volume change and increase of consolidation components were still not readily available, which requires nodule segmentation.
With the real-world clinical workflow and guideline suggestions in mind, we were motivated to develop a CT-based visual forecasting system of lung nodules that can visualize and quantify a nodule in three dimensions (3D) in any future time point. This system can inform the possible growth patterns of a nodule before the actual follow-up scans are taken and help screening personnel to more accurately assess the nodule malignancy and subsequent management. Concretely, we conducted a retrospective study to evaluate the feasibility and performance of our deep learning nodule growth prediction system. We present the following article in accordance with the STARD reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-59/rc).
Methods
Data collection
This retrospective analysis was approved by the Institutional Review Board of Shanghai Chest Hospital (No. KS1956). All procedures performed in this study involving human participants were conducted in accordance with the Declaration of Helsinki (as revised in 2013). Given the retrospective nature of the analysis, the board waived the requirement for patients’ written consent. Anonymity was ensured for all patient data.
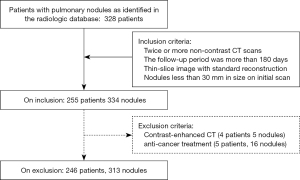
From the radiologic database of our hospital, we consecutively retrieved patients with pulmonary nodules identified on chest CT scans between February 2012 to December 2018. We selected patients for our study using the inclusion and exclusion criteria specified in Figure 1. To be eligible, the patients had to have pulmonary nodules no larger than 30 mm on initial CT scans and have undergone two or more unenhanced thin sliced CT scans using standard algorithm at different time points. We excluded all patients with previously known diagnoses of malignancy treated with chemo/targeted therapy, radiation and surgery (lobectomy/wedge resection). In model development, we excluded all CT images with contrast. We would not exclude patients with contrast enhanced CT scans as long as they had two or more CT scans without contrast.
Image acquisition
Baseline chest CT scans and follow-up examinations were performed using the following 5 scanners: 64-detector row scanner (Brilliance, Philips, Cleveland, OH, USA), 256-detector row scanner (Revolution CT, GE, Waukesha, WI, USA), 128-detector row scanner (Ingenuity core128, Philips, Suzhou, China), Discovery CT750HD CT scanner (GE, Waukesha, WI, USA), and a 16-detector row scanner (uCT S160, United Imaging, Shanghai, China). In the supine position, patients were scanned at the end of inspiration during a single breath hold. The high resolution (HR)CTs were performed with collimation of 0.625–1.25 mm, pitch of 0.64, section thickness of 0.625–1.25 mm without overlap, matrix of 512×512 or 1,024×1,024, field of view (FOV) of 350–400 mm, 120 kVp, and 220–300 mA. All imaging data were reconstructed using the standard algorithm. Follow-up scans were performed to track the therapeutic effectiveness for pneumonia or stability of lung nodules. The mean interval between baseline CT examinations and follow-up scans were 354 days [mean time, 354±224 (SD) days; range, 30 to 1,351 days].
Nodule characterization
The volume of interest (VOI) of the included nodules was manually delineated at voxel level on both baseline and follow-up CT scans by one radiologist (L.Z., with 5 years of experience in chest CT interpretation) using 3D Slicer (version 4.8.0, Brigham and Women’s Hospital, Boston, MA, USA). Then, the VOI was confirmed by another radiologist (X.Y. with 10 years of experience in chest CT interpretation). Large vessels and bronchioles were excluded as much as possible from the volume of the nodules.
Based on the VOIs provided, we computed both geometric properties including mean CT value, diameter, volume, and mass at both baseline and follow-ups, as well as growth properties including VDT and CTR at follow-ups for all included nodules. From the VOI, we got the total number of voxels that a nodule occupies and the CT value in Hounsfield unit (HU) each voxel in the VOI assumes. Then, the volume of a nodule (V in mm3) was computed by multiplying voxel number and the volume of a single voxel; the mean CT value (A in HU) was computed by averaging the CT values of all voxels in the VOI. The mean diameter of a nodule was defined as the average of its maximal length and maximal orthogonal diameter on axial CT slice. Mass in mg was computed as follows: M = V × (A + 1,000)/1,000.
According to the nodule management guidelines, VDT and CTR can be used to triage patients, where above cut-off values indicate high-risk or referable nodules worth further investigation and below cut-off values mean low-risk or non-referable nodule that can be discharged or monitor (6). We computed VDTs using the following equation: [log2× T]/[log(Vfinal/Vinitial)], where Vfinal and Vinitial are the final and initial volumes of the same nodule, respectively, and T is the interval between the final and initial CT scans. Positive VDT indicated possible nodule growth and negative VDT indicated possible nodule shrinkage. We split all the nodules into 2 groups, namely a growth group and non-growth group, based on the relative position of their VDTs compared to cut-off values predefined, where the cut-off for ground glass nodules (GGN, consisting of pure GGN and part-solid nodules) (22) was 1,500 days and that for solid nodule was 500 days. The CTR were computed only for GGN by dividing the nodule volume by the consolidation volume where the nodule regions with a CT value higher than −300 HU were considered as consolidations (23). For CTR, 50% was used as the cut-off.
Nodule growth modelling
To model the nodule growth from baseline CT image to follow-up CT image with a particular time interval, we proposed a convolutional neural network (CNN)-based forecasting system (Figure 2) consisting of two U-Net (24) based networks, the spatial vary network (SVN) and texture vary network (TVN), and one temporal encoding module (TEM). The TEM is developed to encode the time interval between 2 scans into temporal features, which are then incorporated by the SVN and the TVN to learn spatial and texture transformation of a nodule during the interval, respectively. The system is implemented using the PyTorch platform (Meta, Menlo Park, CA, USA) (25) and the details could be found in the supplementary text.

During inference, given the baseline CT image and any time interval as specified by users, the system can predict the imagery representation, both the image and mask, of a pulmonary nodule in the predefined future. In other words, the system treats the image and segmentation mask of a nodule at an initial point as input and then outputs the possible image and segmentation mask of the same nodule at any later time as specified.
To both train and validate the system in a supervised learning fashion, we paired the images and masks at 2 known time points of the same nodule as a data sample. Although the well-trained model could predict the projected image of a nodule in any future time interval, we chose the actual time interval of a follow-up as input to the model so that the model prediction could have ground truth image to be compared with. We pre-processed the data as follows: CT scans are resampled isotropically into 1×1×1 mm3. The voxel intensity (in HU) was first clamped to interval [−1,024, 400] and then normalized to [−1, 1]. Each data sample fed into the model was a cubic volume image with a nodule in the center and had the size of 48×48×48 mm3, which covers the size of all nodules in our study. Due to the limitation of dataset size, we discretized the follow-up time interval using the time mapping function , for most of the intervals are less than 600 days.
Statistical analysis
We split our dataset randomly into 5 groups on patient-level and performed 5-fold cross validation to evaluate our models. That is, all nodules of 1 patient must be in the same subset. The cross validation in our study was nested as the training of our model does not involve feature selection or parameter tuning and the validation data was invisible to our model during training (26). We evaluated the performance of our proposed model in 3 ways. First, we evaluated the CT value, diameter, volume, and mass of the predicted nodule against ground truth using linear regression. Second, we tested the utilities of model predictions in the context of nodule management. As the system outputs both the image and mask of a nodule in a specified future point, we could compute the VDT and CTR. With the VDT and CTR computed from the actual follow-up CT scans, we evaluated how well the model performed in classifying nodule as high-risk versus low-risk by drawing receiver operating characteristic (ROC) curves of the prediction-derived VDT or CTR. Some patients may have many follow-up pairs compared to others, so we only selected the first and last examination of each nodule for classification to eliminate statistical bias.
Quantitative valuables were described as the mean ± standard error, whereas categorical data were presented as numbers (percentages). Clinical and radiologic characteristics were analyzed for differences between the growth and non-growth groups using independent sample t-test for continuous variables and Pearson’s chi-squared test for categorical variables. A 2-sided P<0.05 was considered statistically significant. All analyses were performed using python 3.6 with scikit-learn (v.0.22; https://pypi.org/project/scikit-learn/), SciPy (v.1.3.0; https://pypi.org/project/scipy/), and researchpy (v.0.1.8; https://pypi.org/project/researchpy/).
Results
Patient and nodule characteristics
A total of 313 nodules (115 GGN and 198 solid nodules) from 246 patients (age, 58.7±10.6 years; range, 23 to 97 years) were included in the study. The study cohort consisted of 114 men (age, 59.0±11.2 years; range, 23 to 97 years) and 132 women (age, 58.5±10.2 years; range, 32 to 84 years) and were followed up for an average of 354 days with 3 CT scans on average (range, 2 to 11 scans). The detailed characteristics of the study cohort are displayed in Table 1.
Table 1
| Characteristics | Statistics |
|---|---|
| Age (years) | 58.7±10.6 |
| No. of patients | 246 |
| Female, n (%) | 114 (46.3) |
| Male, n (%) | 132 (53.7) |
| No. of nodules, n (%) | |
| Solitary | 192 (78.0) |
| Multiple | 54 (22.0) |
| Scanning features | |
| CT scanning times | 3 [2–11] |
| Follow-up period (days) | 354±224 |
CT, computed tomography.
The Characteristics of GGNs and solid nodules analyzed in this study is summarized in Tables 2,3. For both GGN and solid nodules, the two groups of different growing patterns differed significantly in VDTs and almost all geometric properties, including mean CT value, diameter, volume, and mass at baseline scans (P<0.05). Besides, the CTR was also significantly different for the 2 growing groups for GGNs.
Table 2
| Characteristics | GGNs (n =115) | Growth group (n=24) | Non-growth group (n=91) | P value of t-test |
|---|---|---|---|---|
| Initial CT value (HU) | −436 (−943 to −197) | −386±66 | −476±79 | <0.001 |
| Initial diameter (mm) | 10.9 (3.0 to 29.8) | 11.9±5.5 | 9.8±4.8 | <0.01 |
| Initial volume (mm3) | 394.0 (27.0 to 7,531.1) | 447.0±274.5 | 394.0±245.1 | <0.001 |
| Initial mass (mg) | 62.6 (5.2 to 5,129.3) | 95.6±146 | 85.6±115 | <0.01 |
| VDT (days) | 1,743 (−40 to 2,305) | 763±548 | 1,805±316 | <0.001 |
| Consolidation volume | 100.86 (0 to 695.4) | 127.84±87.3 | 89.43±74.3 | <0.01 |
| Initial CTR (%) | 25.6 (0.0 to 83.8) | 28.6±23.4 | 22.7±18.6 | <0.01 |
GGN, ground glass nodule; CT, computed tomography; VDT, volume doubling time; CTR, consolidation-to-tumor ratio.
Table 3
| Characteristics | Solid nodule (n=198) | Growth group (n=49) | Non-growth group (n=149) | P value of t-test |
|---|---|---|---|---|
| Initial CT value (HU) | −145 (−347 to −74) | −142±56 | −149±73 | 0.29 |
| Initial diameter (mm) | 8.1 (5 to 28.4) | 8.6±5.3 | 7.3±6.2 | 0.02 |
| Initial volume (mm3) | 171.2 (38.3 to 5,447.2) | 182.6±163.4 | 159.7±155.3 | 0.01 |
| Initial mass (mg) | 128.0 (7.9 to 8,309.2) | 153.2±115.2 | 131.2±109.3 | 0.03 |
| VDT (days) | 543 (−30 to 1,034) | 271±223.2 | 708±189.2, −20 (below 0) | <0.001 |
| Initial CTR (%) | 100 | 100 | 100 | NA |
CT, computed tomography; VDT, volume doubling time; CTR, consolidation-to-tumor ratio.
Model evaluation: geometric properties
We plotted the 4 metrics, including mean CT value, diameter, volume, and mass of the nodules predicted by our system with follow-up time intervals, against that of the nodules on actual follow-up CT scans. As shown by Figure 3, the prediction-derived properties were highly correlated with the real properties (P<0.001) with small relative errors. Compared with the ground truth, our forecasting system generated mean relative errors of 0.083 HU, 0.117 mm, 0.351 mm3, 0.293 mg and for CT value, diameter, volume, and mass, respectively.

Model evaluation: growth properties
Based on the actual CT scans, 24 out of 115 GGNs were high-risk with a VDT larger than 1,500 and 49 out of 198 solid nodules were high-risk with a VDT larger than 500. Using the same cut-off on the VDT derived from our forecasting system, the sensitivity and specificity in distinguishing high-risk from the low-risk were 75.0% and 80.2% for GGNs and 73.5% and 86.6% for solid nodules, respectively. The results have been visualized in Figure 4. Moreover, the predicted VDT by our system had an AUC of 0.857, 0.843, and 0.848 for solid nodules, GGNs, and both types of nodules combined, respectively, in making the risk classification with the actual VDT based grouping as ground truth (Figure 5).


Other than VDT, we also computed the CTR based on the mask and image of GGNs in their final CT scans. With 50% as cut-off value for the CTR, 26 out of 115 GGNs were considered as high-risk nodules (Figure 6A). Our forecasting system had a sensitivity of 0.731, a specificity of 0.876 (Figure 6B), and an AUC of 0.892 (Figure 6C) in making the classification using CTR.

Model evaluation: qualitative assessment
To give an indication of the performance of our forecasting system, we selected 4 nodules with different morphological types and different growth patterns and plotted both their images and masks at baseline scan and at the final scan together with those predicted by our forecasting system (Figure 7). As shown, our forecasting system could make predictions for both solid nodules and GGNs regardless of their real growth patterns, either fast-growing, slow-growing, stable, or shrinkage. Also, the TVN indeed improved the prediction by a single SVN, which is especially evident for the case c in Figure 7.

Discussion
In this proof-of-concept study, we illustrated a deep learning prediction system for lung nodule visualization and growth quantification in any future time point using CTs in hopes of improving the current workflow of LCS instead of just providing a stand-alone score, weakly interpretable to health workers. We evaluated the proposed forecasting system using both geometric and growth properties of lung nodules, demonstrating that the system could accurately forecast the imagery of a nodule in a given future for both GGNs and solid nodules. Although the performance is far from being sufficiently high for the system to be eligible for clinical usage, this study took the first step in the direction of predicting the future imagery of a nodule and laid the foundation for further investigation.
Major efforts have been taken to predict the malignancy of lung nodules either found accidently or identified in the screening settings based solely on initial CT scans. Conventional research has focused on uncovering which radiological signs, such as large size, attachment to vessel, or poor border definition (27) predefined by experts, are associated with or predictive of malignancy. Recent studies have built neural networks under the supervised learning framework to automatically learn representational features that are discriminative for nodule malignancy (13,28). Regardless, these studies have indicated that the imaging snapshot of a nodule contains features informative of its malignancy. While previous studies (13,27,28) have made use of these features only to make binary classifications, this study went a step further by harnessing these informative features to model the projection from a nodule at one time to the same nodule at any later or future times. As shown by our results, this visual modeling of lung nodule growth is feasible and worthy of future studies. A recent study similar to ours built a deep learning model to predict spatial change of lung tumors to facilitate adaptive radiotherapy and achieved good results (29).
In the management of lung nodules, screening or incidental, surveillance or monitoring by follow-up CT scans is a major avenue. Studies have shown that many more follow-up CT scans were performed than baseline CT scans in several national trails of LCS, including NLST (30) and NELSON (31). Machine learning has been successful in predicting lung nodule malignancy based solely on baseline or single CT scans, but it has not incorporated the vast amount of follow-up CT scans or did not take advantage of the consecutive scans of an individual patient to further boost their performance By characterizing the growth pattern of lung nodules, follow-up CT scans could substantially reduce the false positives identified in the baseline CTs (32) and thus help to avoid unnecessary continual follow-up CT scans or invasive procedures. This characterization of growth pattern, as shown by this study, can be modelled by CNN through making full use of the CT scans, both initial and follow-up, collected in LCS. With a much larger dataset than that in this study, it would be promising to enhance the prediction system as a virtual follow-up CT scan specifically designed for lung nodules. This virtual follow-up may even save patients from undergoing unnecessary follow-up CT scans if clinically validated.
As our forecasting system is first of its kind in predicting the visual constituent and mask of a lung nodules in any future point, we explored ways to validate its performance. Based on geometric properties like CT value, diameter, volume, and mass of lung nodules, our system was fairly accurate in depicting the future of a lung nodules in terms of both size and composition. More importantly, we evaluated our system’s ability in discerning on how likely the nodules in question will be malignant and thus require further investigation, using growth properties including VDT and CTR. The VDT, a shape indicator of nodule growth, was found to be significantly associated with nodule malignancy and refined cut-offs by model analysis could reduce false positive by a large margin (5). That is also why we divided the nodules into growth and non-growth groups by thresholding VDT. Tumor composition is as important as size in characterizing lung nodules. The increasing CTR, an index for tumor composition, in GGN has been found to be associated with worse survival for high-risk patients (10). The development of solid components in GGN was also regarded as growth in a study on the natural history of GGNs (33). It is therefore reasonable for us to use CTR as a possible indictor of nodule malignancy to test our system’s performance. As these measures are clinically relevant, the evaluation based on them also demonstrates the usefulness of our forecasting system.
This study had several limitations. First, the study only included a small number of nodules with limited representative capacities and the forecasting system built from this sample may not generalize well in the real-world scenarios. Lung nodules in the real world vary greatly in numerous ways, including shape, size, composition, location, surroundings, as well as growing patterns. A sizable dataset is thus essential for building a forecasting system that can extrapolate the future appearance of heterogeneous lung nodules with decent accuracy. Second, we only explored limited ways in evaluating such forecasting system, including simple geometric and growth measures. Future studies may benefit from incorporating more methods for evaluation, such as comparing HU value distribution between ground truth and prediction within nodule VOI and checking how well radiologists can distinguish between ground truth and prediction visually. Comprehensive evaluation methods are necessary to pave the way for such a forecasting system to enter clinical workflow. Third, we used VDT-derived values to categorize lung nodules into growth or non-growth groups, which are not as perfectly correlated with malignancy as biopsy-based diagnosis. The goal of both actual VDT and model prediction-based VDT was to distinguish between benign and malignant nodules. Thus, it may be more direct to use biopsy-based diagnosis as grouping criteria and ground truth in evaluating the model’s prediction.
To conclude, this pilot study demonstrated that a visual forecasting system of lung nodules based on follow-up CT scans to help health workers manage lung nodules more efficiently and intelligently was fairly accurate when training on a small dataset. After validation with a larger dataset in future studies and integration with automatic segmentation, this forecasting system has the potential to become an automatic virtual follow-up CT system for lung nodules.
Acknowledgments
The authors appreciate the academic support from the AME Radiology Collaborative Group.
Funding: This work was supported by grants from the National Natural Science Foundations of China (Nos. 82071873, 81871353, and 82071990); Shanghai Municipal Health Commission Project (Nos. 2019SY063, 20204Y0201, and 201940221); Shanghai Key Laboratory Open Project (No. STCSM18DZ2270700); Science and Technology Commission Shanghai Municipal project (No. 19411965200); 111 project (No. BP0719010); Xuhui District Artificial Intelligence Medical Hospital Cooperation Project (No. 2020-010); Shanghai “Rising Stars of Medical Talents” Youth Development Program [No. SHWSRS(2021)099]; and China International Medical Foundation (No. Z-2014-07-2003-20).
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-59/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-59/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-59/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This retrospective analysis was approved by the Institutional Review Board of Shanghai Chest Hospital (No. KS1956). All procedures performed in this study involving human participants were conducted in accordance with the Declaration of Helsinki (as revised in 2013). Given the retrospective nature of the analysis, the board waived the requirement for patients’ written consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Oudkerk M, Liu S, Heuvelmans MA, et al. Lung cancer LDCT screening and mortality reduction - evidence, pitfalls and future perspectives. Nat Rev Clin Oncol 2021;18:135-51. [Crossref] [PubMed]
- Ma J, Ward EM, Smith R, et al. Annual number of lung cancer deaths potentially avertable by screening in the United States. Cancer 2013;119:1381-5. [Crossref] [PubMed]
- Gesthalter YB, Koppelman E, Bolton R, et al. Evaluations of Implementation at Early-Adopting Lung Cancer Screening Programs: Lessons Learned. Chest 2017;152:70-80. [Crossref] [PubMed]
- Triplette M, Kross EK, Mann BA, et al. An Assessment of Primary Care and Pulmonary Provider Perspectives on Lung Cancer Screening. Ann Am Thorac Soc 2018;15:69-75. [Crossref] [PubMed]
- Heuvelmans MA, Oudkerk M, de Bock GH, et al. Optimisation of volume-doubling time cutoff for fast-growing lung nodules in CT lung cancer screening reduces false-positive referrals. Eur Radiol 2013;23:1836-45. [Crossref] [PubMed]
- Ruparel M, Quaife SL, Navani N, et al. Pulmonary nodules and CT screening: the past, present and future. Thorax 2016;71:367-75. [Crossref] [PubMed]
- Ye T, Deng L, Wang S, et al. Lung Adenocarcinomas Manifesting as Radiological Part-Solid Nodules Define a Special Clinical Subtype. J Thorac Oncol 2019;14:617-27. [Crossref] [PubMed]
- Baldwin DR, Callister MEGuideline Development Group. The British Thoracic Society guidelines on the investigation and management of pulmonary nodules. Thorax 2015;70:794-8. [Crossref] [PubMed]
- Song YS, Park CM, Park SJ, et al. Volume and mass doubling times of persistent pulmonary subsolid nodules detected in patients without known malignancy. Radiology 2014;273:276-84. [Crossref] [PubMed]
- Matsunaga T, Suzuki K, Takamochi K, et al. What is the radiological definition of part-solid tumour in lung cancer?†. Eur J Cardiothorac Surg 2017;51:242-7. [Crossref] [PubMed]
- Zhang G, Jiang S, Yang Z, et al. Automatic nodule detection for lung cancer in CT images: A review. Comput Biol Med 2018;103:287-300. [Crossref] [PubMed]
- Tao G, Yin L, Shi D, et al. Dependence of radiomic features on pixel size affects the diagnostic performance of radiomic signature for the invasiveness of pulmonary ground-glass nodule. Br J Radiol 2021;94:20200089. [Crossref] [PubMed]
- Bi WL, Hosny A, Schabath MB, et al. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J Clin 2019;69:127-57. [Crossref] [PubMed]
- Causey JL, Zhang J, Ma S, et al. Highly accurate model for prediction of lung nodule malignancy with CT scans. Sci Rep 2018;8:9286. [Crossref] [PubMed]
- da Nóbrega RVM, Rebouças Filho PP, Rodrigues MB, et al. Lung nodule malignancy classification in chest computed tomography images using transfer learning and convolutional neural networks. Neural Comput Appl 2020;32:11065-82. [Crossref]
- Xie Y, Xia Y, Zhang J, et al. Knowledge-based Collaborative Deep Learning for Benign-Malignant Lung Nodule Classification on Chest CT. IEEE Trans Med Imaging 2019;38:991-1004. [Crossref] [PubMed]
- Balagurunathan Y, Beers A, Mcnitt-Gray M, et al. Lung Nodule Malignancy Prediction in Sequential CT Scans: Summary of ISBI 2018 Challenge. IEEE Trans Med Imaging 2021;40:3748-61. [Crossref] [PubMed]
- Gao R, Tang Y, Xu K, et al. Time-distanced gates in long short-term memory networks. Med Image Anal 2020;65:101785. [Crossref] [PubMed]
- Balagurunathan Y, Beers A, Kalpathy-Cramer J, et al. Semi-automated pulmonary nodule interval segmentation using the NLST data. Med Phys 2018;45:1093-107. [Crossref] [PubMed]
- Shi Z, Deng J, She Y, et al. Quantitative features can predict further growth of persistent pure ground-glass nodule. Quant Imaging Med Surg 2019;9:283-91. [Crossref] [PubMed]
- Sheng J, Li Y, Cao G, et al. Modeling nodule growth via spatial transformation for follow-up prediction and diagnosis. In: 2021 International Joint Conference on Neural Networks (IJCNN); 2021:1-7.
- Goo JM, Park CM, Lee HJ. Ground-glass nodules on chest CT as imaging biomarkers in the management of lung adenocarcinoma. AJR Am J Roentgenol 2011;196:533-43. [Crossref] [PubMed]
- Shimomura M, Iwasaki M, Ishihara S, et al. Volume-Based Consolidation-to-Tumor Ratio Is a Useful Predictor for Postoperative Upstaging in Stage I and II Lung Adenocarcinomas. Thorac Cardiovasc Surg 2019; [Crossref] [PubMed]
- Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N, Hornegger J, Wells WM, et al. editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Cham: Springer International Publishing; 2015:234-41.
- Paszke A, Gross S, Chintala S, et al. Automatic differentiation in PyTorch. 2017 Oct 28 [cited 2021 Jul 18]; Available online: https://openreview.net/forum?id=BJJsrmfCZ
- Vabalas A, Gowen E, Poliakoff E, et al. Machine learning algorithm validation with a limited sample size. PLoS One 2019;14:e0224365. [Crossref] [PubMed]
- Liu Y, Wang H, Li Q, et al. Radiologic Features of Small Pulmonary Nodules and Lung Cancer Risk in the National Lung Screening Trial: A Nested Case-Control Study. Radiology 2018;286:298-306. [Crossref] [PubMed]
- Ardila D, Kiraly AP, Bharadwaj S, et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med 2019;25:954-61. [Crossref] [PubMed]
- Wang C, Rimner A, Hu YC, et al. Toward predicting the evolution of lung tumors during radiotherapy observed on a longitudinal MR imaging study via a deep learning algorithm. Med Phys 2019;46:4699-707. [Crossref] [PubMed]
- National Lung Screening Trial Research Team. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409. [Crossref] [PubMed]
- Horeweg N, van der Aalst CM, Vliegenthart R, et al. Volumetric computed tomography screening for lung cancer: three rounds of the NELSON trial. Eur Respir J 2013;42:1659-67. [Crossref] [PubMed]
- Pinsky PF, Gierada DS, Black W, et al. Performance of Lung-RADS in the National Lung Screening Trial: a retrospective assessment. Ann Intern Med 2015;162:485-91. [Crossref] [PubMed]
- Qi LL, Wu BT, Tang W, et al. Long-term follow-up of persistent pulmonary pure ground-glass nodules with deep learning-assisted nodule segmentation. Eur Radiol 2020;30:744-55. [Crossref] [PubMed]