
Clinicopathologic characteristics and diagnostic methods of RET rearrangement in Chinese non-small cell lung cancer patients
Introduction
The discovery of targetable oncogenic drivers has led to significant improvements in the treatment of non-small cell lung cancer (NSCLC). Among these, a number of fusion drivers, such as anaplastic lymphoma kinase (ALK), c-ros oncogene 1 (ROS1), and rearranged during transfection (RET), have been described as rare oncogenic events and recommended for routine testing at diagnosis (1).
The RET proto-oncogene was first identified in 1985 by Takahashi et al. (2). It encodes a transmembrane receptor tyrosine kinase that plays a key role in the differentiation of the kidneys and nervous system (3,4). The rearrangement of RET can lead to the constitutive activation of the RET tyrosine kinase domain and the recruitment of downstream signaling cascades, such as MAP kinase (MAPK), phosphatidylinositol-3-kinase (PI3K)/Akt, and Janus kinase-signal transducer and activator of transcription (JAK-STAT) pathways, which promote tumorigenesis (5).
Several multi-kinase inhibitors (MKIs), such as vandetanib and cabozantinib, have been used for targeted therapy to treat advanced RET-rearranged NSCLC patients. However, MKIs sometimes leads to significant “off-target” side effects, such as nausea, diarrhea, and hypertension (6,7). Recently, novel RET tyrosine kinase inhibitors (TKIs) with high selectivity, including selpercatinib (LOXO-292) and pralsetinib (BLU-667), have been approved by the Food and Drug Administration (FDA) for the treatment of advanced RET-rearranged lung and thyroid cancer (8-10). Therefore, it is important to rapidly and accurately identify RET-positive cases.
However, due to the low prevalence of RET fusions, there is only limited information about the clinical characteristics and outcomes of RET-rearranged NSCLC, especially in Chinese patients. Incidence of RET rearrangement ranges from 1% to 2% in NSCLC (11-13), and depends on the age, sex, smoking history and histological subtype. Different fusion partners have been identified in NSCLC, with the most major proportion being KIF5B (14). RET rearrangements tend to be mutually exclusive with other driver mutations in NSCLC such as EGFR, KRAS, ALK and ROS1, and associate with low PD-L1 expression (15) and low tumor mutation burden (16).
Furthermore, the screening methods for RET rearrangement have not yet been standardized. Several molecular diagnostic methods are used to identify gene fusions, including IHC, RT-PCR, FISH, and DNA/RNA-based NGS. Although IHC is an effective screening tool to detect ALK-positive patients, it showed low sensitivity (55–65%) and variable specificity (40–85%) in prior study (17) and may not be reliable to detect RET rearrangement. RT-PCR is specific, but it is limited to known fusion partners and thus may underestimate prevalence. RET FISH is highly sensitive (100%) but has suboptimal specificity (45–60%) (17,18). Moreover, break-apart displays low sensitivity in detecting non-canonical RET fusions (19). DNA-based NGS enables the detection of high-throughput genomic alterations and novel partners of gene rearrangement, but it fails to provide information on functional fusion transcripts (20). The main laboratory methods for RET rearrangements have not been systematically investigated and fully elucidated in NSCLC. Therefore, investigation of the prevalence, characteristics, and diagnostic methods in a large cohort of NSCLC patients may provide comprehensive genomic profiling and optimal strategy for the selection of RET-rearranged patients.
This study analyzed the molecular profile of 9,431 Chinese NSCLC patients who underwent targeted DNA-based NGS in daily clinical practice at two large cancer centers and identified 167 RET-rearranged NSCLC cases. Different techniques, including RNA-based NGS, FISH, and IHC, were performed to investigate their performance. The prevalence, clinicopathologic characteristics, molecular profiling, and therapeutic outcomes of the RET-rearranged cases were analyzed. We present the following article in accordance with the REMARK reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-202/rc).
Methods
Patients and study design
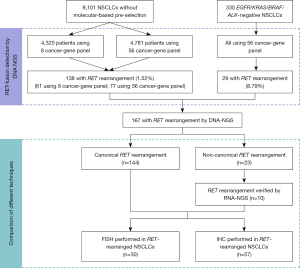
This retrospective analysis included 9,431 NSCLC patients in a multi-center study from January 2017 to August 2020 (including the Henan Cancer Hospital, Zhengzhou, China and National Cancer Center/Cancer Hospital, Chinese Academy of Medical Science and Peking Union Medical College, Beijing, China). All patients who met the following criteria were included in the analysis: confirmed NSCLC by pathology; detected the mutations by NGS (8/56 cancer-related genes). Altogether, the cohort included 9,101 NSCLC patients without mutation-based pre-selection and 330 epidermal growth factor receptor (EGFR)/KRAS proto-oncogene (KRAS)/B-Raf proto-oncogene (BRAF)/ALK-negative patients (EGFR/KRAS/BRAF mutation status was tested by PCR, and ALK fusion status was tested by IHC-Ventana) (Figure 1). RET-positive cases were collected and analyzed. RNA-NGS were performed in non-canonical fusion subtypes. FISH (N=30) and IHC (N=57) assays were performed in RET-rearranged patients with sufficient tissue. Patients’ medical records were retrospectively reviewed to collect data on age, sex, smoking status, tumor stage at diagnosis, pathological diagnosis, and treatment histories. The stage of each patient was assessed following American Joint Committee on Cancer Staging Manual version 7. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the local ethics committee of The Affiliated Cancer Hospital of Zhengzhou University (No. 2021-KY-0092), and was also approved by the institutional review board of National Cancer Center/Cancer Hospital, Chinese Academy of Medical Science and Peking Union Medical College (No. 20/444-2640). Written informed consent was obtained from all individuals included in the study.
Targeted DNA-NGS
Genomic DNA from formalin-fixed paraffin embedded (FFPE) tissue was isolated using the QIAamp DNA FFPE Tissue Kit (Qiagen, Duesseldorf, Germany). Libraries were prepared using commercial panels (Burning rock Technology, Guangzhou, China). Briefly, a panel covering 8 driver genes of NSCLC was used on 4,320 patients from the Henan Cancer Hospital. A panel covering 56 cancer-related genes was used on the other patients enrolled in the study, as previously reported (21). Samples were sequenced on the Nextseq 550 (Illumina, San Diego, CA, USA) with an average depth of 1,000×. All the reads were mapped to human genome 19, and alignments were visualized using the Integrative Genomics Viewer.
Targeted RNA-NGS
Total RNA from the FFPE tissue was extracted using the Magen FFPE DNA/RNA kit (Magen, Guangzhou, China). The quantity and quality of RNA were detected using the Qubit RNA HS Assay Kit (Thermo Fisher Scientific) and the RNA Pico Sensitivity Reagent Kit (PerkinElmer), respectively. Libraries were prepared using the commercial panel (Burning rock Technology) containing 115 fusions and splice-region variants, which covered the entire coding region of RET. NGS was performed on the Novaseq-6000 platform (Illumina) with at least 25M reads per sample.
FISH and IHC
FISH was performed using the RET (10q11) dual-color break-apart rearrangement probe (LBP Medicine Science and Technology, Guangzhou) and following the manufacturer’s instructions. For each sample, >100 tumor cells were evaluated. Samples were considered RET-rearrangement when ≥15% of the tumor cells showed split signals or isolated 3' signals. Isolated 5' signals were thought to result from the deletion of the kinase domain and were considered negative. IHC was performed using a rabbit monoclonal anti-RET (EPR2871) antibody (ab134100, Abcam, Cambridge, MA). RET expression was evaluated according to the following intensity scores: 0: negative; 1+: weak; 2+: moderate, and 3+: strong in >10% of tumor cells. The FISH and IHC results were evaluated by 2 pathologists independently, blinded to the NGS results. We evaluated the RET-fusion using NGS as the reference standard in this study. FISH and IHC assays were performed in RET-rearranged patients with sufficient tissue. Consistency was defined as the percent of positive events to NGS results.
Statistical analysis
The Kaplan-Meier method was used to determine progression-free survival (PFS), and differences between groups were calculated using the log-rank test. The treatment response was assessed according to the Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1 (22). All the statistical analyses were conducted using GraphPad Prism 5 software. The clinical characteristics of the different groups, including gender, age, smoking history, histology, and brain metastasis, were compared by the χ2 test or Fisher exact test. Cox’s proportional-hazards model was used to estimate the hazards ratio (HR) and the corresponding 95% confidence interval (95% CI) for the covariates of interests. Variables included sex, smoking, age, histology, stage, RET-rearranged subtype, breakpoint, distant metastasis, brain metastasis were selected for univariate analysis. Covariates with P value <0.10 from univariate analysis were considered for multivariable model. Statistical significance was defined as a 2-sided P value <0.05.
Results
Prevalence and characteristics of RET-rearranged NSCLC
In total, 167 RET-rearranged NSCLC patients were detected in our study. Among the 9,101 NSCLC patients without mutation-based pre-selection, 1.52% (138/9,101) were detected to have RET rearrangement, while in EGFR/KRAS/BRAF/ALK-negative NSCLC patients, the prevalence of RET rearrangement was 8.79% (29/330) (Figure 1).
The clinicopathological characteristics were accessible for 129 RET-rearranged NSCLC patients (Table 1). The median age at diagnosis was 57 years (range, 30–83 years). RET rearrangements were more frequent in females (65.1%) and never smokers (82.2%). The most common histological subtype of RET-rearranged NSCLC patients was lung adenocarcinoma (92.3%), but other subtypes were also detected, including squamous cell carcinoma (1.5%), large cell carcinoma (0.8%), and neuroendocrine carcinoma (1.5%). Among the 62 stage IV RET-rearranged patients, 74.2% (46/62) had distant metastasis, such as bone, brain, or liver metastasis. Notably, 40.3% (25/62) of the stage IV RET-rearranged patients had brain metastasis.
Table 1
| Patient characteristics | No. (%) of patients (N=129) |
|---|---|
| Median age, years [range] | 57 [30–83] |
| Gender | |
| Female | 84 (65.1) |
| Male | 45 (34.9) |
| Smoking | |
| Never | 106 (82.2) |
| Smoker | 23 (17.8) |
| Histology | |
| Adenocarcinoma | 119 (92.3) |
| Squamous | 2 (1.5) |
| Adenosquamous | 5 (3.9) |
| Large cell carcinoma | 1 (0.8) |
| Neuroendocrine carcinoma | 2 (1.5) |
| Ki-67 (%) | |
| <20% | 9 (7.0) |
| 20–39% | 13 (10.1) |
| 40–59% | 9 (7.0) |
| ≥60% | 18 (13.9) |
| NA | 80 (62.0) |
| Stage at diagnosis | |
| I | 37 (28.7) |
| II | 7 (5.4) |
| III | 23 (17.8) |
| IV | 62 (48.1) |
| Distant metastasis (% of stage IV) | |
| No | 16 (25.8) |
| Yes | 46 (74.2) |
| Brain metastasis (% of stage IV) | |
| No | 37 (59.7) |
| Yes | 25 (40.3) |
RET, rearranged during transfection; NSCLC, non-small cell lung cancer; NA, not available.
Partners of RET fusion
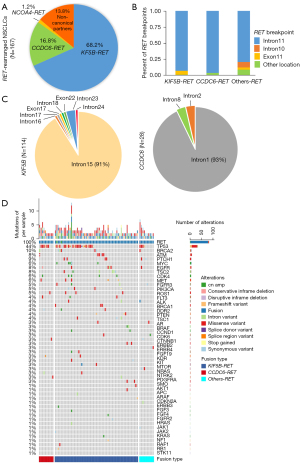
Among the 167 RET rearrangements, the most common fusion partner was KIF5B (68.2%, 114/167), followed by coiled-coil domain containing 6 (CCDC6) (16.8%, 28/167), and nuclear receptor coactivator 4 (NCOA4) (1.2%, 2/167) (Figure 2A). The breakpoint of RET was most frequently observed in intron 11, but other breakpoints included intron 10 and exon 11 (Figure 2B). In relation to the fusion partners, the most common breakpoints were KIF5B intron 15 (91% of KIF5B-RET patients) and CCDC6 intron 1 (93% of CCDC6-RET patients) (Figure 2C).
Different characteristics were compared between the 90 KIF5B-RET patients and 23 CCDC6-RET patients with accessible clinical records (Table S1). The incidence of brain metastasis and distant metastasis in KIF5B-RET patients was higher than that in CCDC6-RET patients; however, the differences were not statistically significant.
Additionally, non-canonical fusion partners were also identified in 23 patients, including ADAM metallopeptidase with thrombospondin type 1 motif 2 (ADAMTS2), Rho GTPase activating protein 12 (ARHGAP12), centrosomal protein 128 (CEP128), epidermal growth factor receptor pathway substrate 8 (EPS8), and intergenic fusions. The relevant clinical information is listed in Table 2. Of the fusion partners, 3 have been reported as individual cases in the literature (23-25), but others have never been reported. RNA-based NGS was then performed to verify non-canonical RET rearrangement. Among the 23 non-canonical fusion cases, 10 cases had samples available for RNA-NGS and were all proven to have functional RET fusions at the RNA level (Table 2). It is of great clinical significance to evaluate the RET-TKI efficacy in such cases.
Table 2
| Fusion | Breakpoints | Locus of the partner gene | Gender | Age (years) | Histology | Literature | RNA-NGS results | FISH | IHC |
|---|---|---|---|---|---|---|---|---|---|
| ADAMTS2-RET | Intron10_Exon3 | 5q35.3 | M | 54 | ADC | NA | NA | NA | NA |
| ARHGAP12-RET | Intron4_Intron11 | 10p11.22 | F | 65 | ADC | NA | KIF5B-RET (Exon15_Exon12) | + | 1+ |
| CEP128-RET | Intron18_Intron10 | 14q31.1 | F | 63 | ADC | NA | NA | NA | NA |
| EPS8-RET | Intron12_Intron11 | 12p12.3 | F | 56 | ADC | NA | EPS8-RET (Exon12_Exon12) | NA | 1+ |
| ERC1-RET | Intron8_Intron11 | 12p13.33 | M | 54 | ADC | PMID: 32737449 | NA | NA | − |
| KIAA1217-RET | Intron1_Intron11 | 10p12.2-p12.1 | F | 70 | ADC | PMID: 31162284 | KIF5B-RET (Exon15_Exon12) | + | 2+ |
| PLCXD3-RET/LINC01264-RET | Intron2_Intron11/intergenic_Intron11 | 5p13.1; 10q11.21 | M | 83 | ADC | NA | KIF5B-RET (Exon15_Exon12) | NA | NA |
| SLC6A11-RET | Intron5_Intron11 | 3p25.3 | M | 71 | ADC | NA | NA | NA | NA |
| SPECC1L-DORA2A-RET | Intron10_Intron11 | 22q11.23 | F | 60 | ADC | PMID: 31917708 | NA | NA | NA |
| STK33-RET | Intron1_Intron11 | 11p15.4 | F | 65 | ADC | NA | NA | NA | NA |
| BET1-RET | Intergenic_Intron11 | 7q21.3 | F | 56 | ADC | NA | KIF5B-RET (Exon15_Exon12) | + | 1+ |
| CENPK-RET | Intergenic_Exon12 | 5q12.3 | M | 58 | SCC | NA | NA | NA | NA |
| FXYD4-RET | Intergenic_Intron11 | 10q11.21 | F | 54 | ADC | NA | NA | NA | NA |
| LINC00680-RET | Intergenic_Intron10 | 6p11.2 | F | 63 | ADC | NA | GOLGA5-RET (Exon7_Exon12) | + | 1+ |
| KIAA0146-RET | Intergenic_Intron11 | 8q11.21 | M | 65 | SCC | NA | NA | NA | NA |
| LOC105378330-RET | Intergenic_Intron11 | 10q21.3 | F | 64 | ADC | NA | NA | NA | NA |
| LOC105378470-RET | Intergenic_Intron11 | 10q25.1 | F | 64 | ADC | NA | KIF5B-RET (Exon15_Exon12) | NA | NA |
| LOC441666-RET | Intergenic_Intron11 | 10q11.21 | F | 67 | ADC | NA | NA | NA | NA |
| MARCH8-RET | Intergenic_Intron11 | 10q11.21-q11.22 | F | 56 | ADC | NA | KIF5B-RET (Exon15_Exon12) | + | 2+ |
| NAMPTL-RET | Intergenic_Intron11 | 10p11.21 | F | 49 | AdCa | NA | NA | NA | NA |
| OR13A1-RET | Intergenic_Intron11 | 10q11.21 | M | 58 | ADC | NA | NA | NA | NA |
| TNIP1-RET/ RASGEF1A-RET | Intron8_Intron11/intergenic_Intron11 | 5q33.1; 10q11.21 | M | 65 | ADC | NA | TNIP1-RET (Exon8_Exon12) | − | 3+ |
| TBC1D14-RET | Intergenic_Intron11 | 4p16.1 | F | 65 | ADC | NA | KIF5B-RET (Exon15_Exon12) | NA | NA |
F, female; M, male; ADC, adenocarcinoma; SCC, Squamous Cell Carcinoma; AdCa, Adenosquamous; NA, not available; FISH, fluorescence in situ hybridization; IHC, immunohistochemistry; −, negative; +, positive; 1+, weak; 2+, moderate; 3+, strong.
Mutation profile and concurrent driver gene alterations
As Figure 1 shows, 106 RET-arranged patients were detected using 56 cancer-related gene panel. Co-occurring genetic aberrations were found in 77 patients (77/106, 73%). We constructed a heatmap to demonstrate the alterations co-occurring with the RET rearrangements (Figure 2D). Tumor protein 53 (TP53) was the most commonly altered (34/77, 44%), followed by BRCA2 (8/77, 10%), PTCH1 (7/77, 9%), ATM (6/77, 8%), EGFR (6/77, 8%), and TSC2 (6/77, 8%). Other genomic alterations, including MYC, CDK4, MET, FGFR3, and PIK3CA, were also observed. Among the 106 RET-rearranged patients, 8 (7.55%) harbored concurrent driver gene alterations, including EGFR L858R (N=3), EGFR 19del (N=2), KRAS G12X (N=2), and EML4-ALK (N=1) (for further details, see Table S2).
Treatment and clinical outcomes of RET-rearranged NSCLC
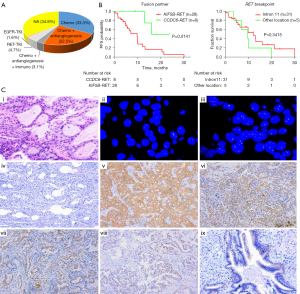
Among the 129 RET-rearranged NSCLC patients with available treatment information, approximately 33.3% (43/129) underwent platinum-based doublet chemotherapy as the first-line treatment, 22.5% (29/129) received chemotherapy combined with antiangiogenic therapy as the first-line treatment, and 3.1% (4/129) received immune checkpoint inhibitors in clinical trials (Figure 3A). To evaluate the chemotherapy efficacy among different subtypes, the survival data of 36 late-stage patients were analyzed, including 28 KIF5B-RET patients and 8 CCDC6-RET patients. We found that patients with the CCDC6-RET subtype had significantly longer PFS than those with the KIF5B-RET subtype (23 vs. 9.7 months; P=0.014) (Figure 3B). Additionally, no significant difference was observed between the different breakpoints of RET (intron 11 vs. other locations) (Figure 3B). As KIF5B-RET patients appeared to suffer from brain and distant metastasis more than CCDC6-RET patients (Table S1; albeit the difference was not statistically significant), RET-rearranged subtypes were included in the Cox proportional-hazards model with other clinical characteristics. The results of the univariate and multivariate Cox proportional-hazards model based on the 36 RET-rearranged cases are listed in Table S3. Covariates with a P value <0.10 in the univariate analysis were included in the multivariable model. According to the multivariable analysis, CCDC6-RET cases had significantly better PFS than KIF5B-RET cases (HR =0.192, 95% CI: 0.044–0.831; P=0.027). Research with a sufficiently large cohort needs to verify these findings.
Only 4.7% (6/129) of the patients had access to RET-TKI, mainly due to the inaccessibility of RET-TKI at that time. A 54-year-old male with poorly differentiated lung adenocarcinoma had disease progression after receiving surgery, radiotherapy, and chemotherapy. The targeted NGS revealed an ERC1-RET fusion. Subsequently, after being started on Cabozantinib, SD was achieved. The patient continued to receive Cabozantinib treatment for 10 months before disease progression with new lung metastasis (Figure S1).
None of the 8 patients with concurrent driver gene alterations received RET-TKI treatment. One lung adenocarcinoma patient (Case No. 1) with EGFR L858R and KIF5B-RET has received Icotinib for almost 2 years and achieved stable disease (SD). When the patient’s disease progressed, KIF5B-RET continued to be detected, but EGFR L858R disappeared. Another lung adenocarcinoma patient with EGFR 19del (Case No. 5) has been receiving Gefitinib for 2 years, and CCDC6-RET was detected as a resistant mechanism in the EGFR-TKI relapsed tumor (Table S2).
Detection of RET rearrangement
Among the 167 RET-positive cases detected by DNA-NGS, 144 were canonical fusion subtypes, and 23 were non-canonical fusion subtypes. A total of 10 non-canonical fusion samples were available for RNA-NGS, and all were proven to have functional RET fusions at the RNA level (Table 2). Representative fusion patterns at DNA and RNA levels are shown in Figure S2 using IGV.
FISH (N=30) and IHC (N=57) assays were performed in RET-rearranged patients with sufficient tissue. The FISH results revealed the RET rearrangement in 25/30 patients, resulting in the FISH/NGS concordance of 83.3% (Table 3). IHC intensity scores were 0 in 3.5% (2/57), 1+ in 35.1% (20/57), 2+ in 33.3% (19/57), and 3+ in 28.1% (16/57) of the RET-rearranged patients. IHC 3+ was considered positive, and the concordance of IHC and NGS was 28.1%. The staining pattern of RET-IHC varied in different fusion subtypes. Notably, KIF5B-RET showed diffuse, 2+/3+ cytoplasmic staining, while CCDC6-RET and others showed granular and patchy staining with weak intensity (Figure 3C). The normal tracheal epithelium also showed RET-IHC staining, which might lead to a staining pitfall in the interpretation of IHC results. We noted that IHC had extremely low sensitivity in non-KIF5B-RET patients (CCDC6-RET, 8%, NCOA4-RET, 0%, and other-RET, 25%), while FISH also showed unsatisfying sensitivity in non-KIF5B-RET patients. FISH or IHC were not able to detect NCOA4-RET cases.
Table 3
| Test result | NGS status | ||||
|---|---|---|---|---|---|
| KIF5B-RET | CCDC6-RET | NCOA4-RET | Others-RET | All patients | |
| RET-IHC | |||||
| N | 39 | 12 | 2 | 4 | 57 |
| 3+ | 14 | 1 | 0 | 1 | 16 |
| 2+ | 14 | 5 | 0 | 0 | 19 |
| 1+ | 11 | 6 | 1 | 2 | 20 |
| 0 | 0 | 0 | 1 | 1 | 2 |
| Concordance | 35.9% | 8.3% | 0.0% | 25.0% | 28.1% |
| RET-FISH | |||||
| N | 19 | 7 | 2 | 2 | 30 |
| Positive | 18 | 6 | 0 | 1 | 25 |
| Negative | 1 | 1 | 2 | 1 | 5 |
| Concordance | 94.7% | 85.7% | 0.0% | 50.0% | 83.3% |
RET, rearranged during transfection; IHC, immunohistochemistry; FISH, fluorescence in situ hybridization; 0, negative; 1+, weak; 2+, moderate; 3+, strong; NGS, next-generation sequencing.
Discussion
To the best of our knowledge, this study has one of the largest RET-rearranged NSCLC cohorts for which a comprehensive analysis of molecular profiling, clinical outcomes, and detection methods has been performed. This study enrolled 9,431 Chinese NSCLC patients, among whom NGS identified 167 RET-rearranged patients. In 9,101 Chinese NSCLC patients without molecular-based pre-selection, the prevalence of RET rearrangement was 1.52%, reflecting the findings of previous reports (26,27). In 330 EGFR/KRAS/BRAF/ALK-negative NSCLC patients, the prevalence of RET rearrangement was up to 8.8%, indicating the necessity of RET detection in NSCLC when other driver genes are negative. Similar to ALK- and ROS1-rearranged NSCLC (28), RET rearrangement was more common in female, never smokers, and lung adenocarcinoma patients. Importantly, 40.3% (25/62) of the stage IV RET-rearranged NSCLC patients had brain metastasis. This is much higher than the average brain metastasis rate reported for advanced NSCLC (10–20%) (29), and is especially high in KIF5B-RET patients (43%). Previous studies have also reported that RET fusion is an independent risk factor of brain metastasis (30,31). This observation may reinforce the importance of evaluating the intracranial therapeutic response of RET-TKIs based on the molecular subclass of tumors. Selective RET inhibitors, including pralsetinib and selpercatinib, have been approved by the FDA, and both of them have shown a significant ability to cross the blood-brain barrier (31-33).
To date, at least 15 RET-rearranged subtypes have been reported in NSCLC, including KIF5B-RET, CCDC6-RET (34), NCOA4-RET (35), TRIM33-RET (36), KIAA1217-RET (37), ERC1-RET (38), and MYO5C-RET (39). In this study, diverse RET fusion partners were identified, including canonical partners, such as KIF5B (68.2%, 114/167), CCDC6 (16.8%, 28/167), and NCOA4 (1.2%, 2/167). Rare partners, such as KIAA1217 and TBC1D32, were also identified, which have been reported in a previous study (40). Among the 23 non-canonical fusion subtypes identified in our study by DNA-NGS, 10 with sufficient tumor tissue were verified to harbor functional fusion transcripts by RNA-NGS, and 3 novel partners (EPS8, GOLGA5, and TNIP1) were found. Numerous breakpoints of ALK rearrangement have been reported to be associated with clinical benefits (17,41). However, the breakpoints of RET and its partners were relatively concentrated in our study, mainly in RET intron 11, KIF5B intron 15, and CCDC6 intron 1 and no significant survival difference for chemotherapy was observed between the different RET breakpoints.
We also characterized the mutational profile of the RET-rearranged patients and found that TP53 was the most common concurrent alteration. Previous studies have suggested that TP53 concomitant mutations have a strong negative effect on the outcomes of patients with EGFR-mutant (42-45) and ALK-rearranged NSCLC (43,46,47). The poor prognostic effect of the TP53 mutation on tumors may be due to the loss of tumor inhibitory function and the elevated level of genomic instability (48). It is generally believed that RET rearrangement occurs exclusively with other oncogenic drivers in treatment-naive lung cancers (49). However, 8 RET-rearranged NSCLC patients in our study harbored concurrent oncogenic driver gene alterations. CCDC6-RET was found to be a resistant mechanism in an EGFR L858R patient, and the 7 other RET-rearranged NSCLC patients with concurrent driver gene alterations were all treatment-naïve patients. Among these treatment-naïve patients, only one KIF5B-RET patient with EGFR L858R received EGFR-TKI Icotinib treatment, and that patient had a PFS time of 23 months. Intratumor heterogeneity may explain multi-driver gene alterations. Sun (50) and Kim (51) reported that RET fusion could occur as an acquired resistance mechanism to Osimertinib. Additionally, McCoach (52) reported that RET rearrangement could also act as an acquired resistance mechanism of ALK-TKI. Thus, we believe that screening for RET fusion in post-treatment settings is clinically significant.
Due to the inaccessibility of the RET-TKI at the time of diagnosis in our study cohort, most RET-rearranged patients received chemotherapy. Pemetrexed-based chemotherapy for NSCLC patients with RET fusion-positive metastatic NSCLC has been shown to provide a durable benefit (53). In our study, the CCDC6-RET subgroup had a significantly longer PFS than the KIF5B-RET subgroup. Tan et al. reported that overall survival was more prolonged in CCDC6-RET fusion than KIF5B-RET fusion-positive patients (54). However, the reasons for a better prognosis in CCDC6-RET than KIF5B-RET patients remain unclear.
Precision medicine for tumors depends on an effective and reliable detection method. This is even more important for mutations which exist only in a small proportion of patients. Ideally, molecular detection should be highly sensitive, specific, and feasible in most diagnostic laboratories. At present, there is no gold-standard for RET rearrangement detection. FISH has been the gold-standard assay for diagnosing ALK- and ROS1-fusions (55). It is available in most pathology laboratories, and has a low tumor tissue requirement. However, our study revealed that RET FISH might lead to false-negative results, especially in CCDC6-RET and non-canonical RET-fusion subtypes. Technically, the RET FISH test may be more challenging than most other break-apart assays, as RET and its most common fusion partners are situated very near to each other on chromosome 10 (approximately 7.9–17.9 Mb apart) and are thus difficult to interpret. Radonic indicated that FISH is a sensitive but unspecific technique for RET screening (56). RET-IHC has not generally been recommended in previous studies, as IHC is more likely to yield false-negative results (17,57,58), which was also observed in our study. The current impediments of RET-IHC include the low-level expression of the RET-fusion protein and the lack of specific antibodies. We also observed RET expression in the normal tracheal epithelium, which might lead to a false-positive result.
Targeted DNA-NGS in RET detection is accurate and comprehensive and thus provides a unique advantage in exploring novel partners and the simultaneous testing of multiple genes. However, DNA-NGS has limitations in identifying complex fusions, and RNA-NGS adds value to accurate detection (Figure S2). The discordance of RET fusion at the DNA and RNA level may be due to alternative splicing and the flexible break-induced repair mechanisms (59,60). Taking all these factors into consideration, we recommend DNA-NGS as a preliminary screening strategy for patients who have been newly diagnosed. A FISH analysis may be an appropriate method when the specimens have too low tumor cell content. For unusual results of DNA-NGS or FISH, RNA-NGS can be used as a validation technique.
There are several limitations of our study. It is a retrospective study, in only two major centers. The NGS methods were not completely the same in the whole study cohort. Another limitation of our study is that due to the unavailability of targeting agents, the number of patients receiving RET-TKIs was small. Therefore, no correlation could be done between the clinical efficacy of RET-TKIs with fusion. We intend to conduct further studies that include more clinical features of RET-rearranged patients and prognostic evaluations of different RET-fusion subtypes.
Acknowledgments
The authors appreciate the academic support from the AME Lung Cancer Collaborative Group. The abstract has been presented at the Chinese Academic Conference on Tumor Biomarker (CCTB 2021).
Funding: This work was supported by the major public welfare projects in Henan Province (grant No. 201300310400) and the Technological and scientific projects in Henan Province (grant Nos. LHGJ20210175 and 222102310136).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-202/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-202/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-202/coif). LB reports that he has received grants from Takeda, Roche, BMS; payment or honoraria for lectures, presentations, speakers bureaus from Amgen, AstraZeneca, BMS, Eli Lilly, Janssen, MSD, Merck, Novartis; participation on an Advisory Boards of Eli Lilly, Janssen, MSD, Merck, Novartis. He is an International Secretary of the Austrian Society of Pathology, Member of Pulmonary Pathology Society Membership and Awards Committee, and Member of IASLC Mesothelioma Committee. TH has received payment for speakers bureaus from Chugai Pharmaceutical, outside the submitted work. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the local ethics committee of The Affiliated Cancer Hospital of Zhengzhou University (No. 2021-KY-0092), and was also approved by the institutional review board of National Cancer Center/Cancer Hospital, Chinese Academy of Medical Science and Peking Union Medical College (No. 20/444-2640). Written informed consent was obtained from all individuals included in the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lindeman NI, Cagle PT, Aisner DL, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Mol Diagn 2018;20:129-59. [Crossref] [PubMed]
- Takahashi M, Ritz J, Cooper GM. Activation of a novel human transforming gene, ret, by DNA rearrangement. Cell 1985;42:581-8. [Crossref] [PubMed]
- Ishizaka Y, Itoh F, Tahira T, et al. Human ret proto-oncogene mapped to chromosome 10q11.2. Oncogene 1989;4:1519-21. [PubMed]
- Arighi E, Borrello MG, Sariola H. RET tyrosine kinase signaling in development and cancer. Cytokine Growth Factor Rev 2005;16:441-67. [Crossref] [PubMed]
- Ibáñez CF. Structure and physiology of the RET receptor tyrosine kinase. Cold Spring Harb Perspect Biol 2013;5:a009134. [Crossref] [PubMed]
- Bronte G, Ulivi P, Verlicchi A, et al. Targeting RET-rearranged non-small-cell lung cancer: future prospects. Lung Cancer (Auckl) 2019;10:27-36. [Crossref] [PubMed]
- Subbiah V, Cote GJ. Advances in Targeting RET-Dependent Cancers. Cancer Discov 2020;10:498-505. [Crossref] [PubMed]
- Bradford D, Larkins E, Mushti SL, et al. FDA Approval Summary: Selpercatinib for the Treatment of Lung and Thyroid Cancers with RET Gene Mutations or Fusions. Clin Cancer Res 2021;27:2130-5. [Crossref] [PubMed]
- Subbiah V, Shen T, Terzyan SS, et al. Structural basis of acquired resistance to selpercatinib and pralsetinib mediated by non-gatekeeper RET mutations. Ann Oncol 2021;32:261-8. [Crossref] [PubMed]
- Markham A. Pralsetinib: First Approval. Drugs 2020;80:1865-70. [Crossref] [PubMed]
- Li AY, McCusker MG, Russo A, et al. RET fusions in solid tumors. Cancer Treat Rev 2019;81:101911. [Crossref] [PubMed]
- Takeuchi S, Yanagitani N, Seto T, et al. Phase 1/2 study of alectinib in RET-rearranged previously-treated non-small cell lung cancer (ALL-RET). Transl Lung Cancer Res 2021;10:314-25. [Crossref] [PubMed]
- Zhang K, Chen H, Wang Y, et al. Clinical Characteristics and Molecular Patterns of RET-Rearranged Lung Cancer in Chinese Patients. Oncol Res 2019;27:575-82. [Crossref] [PubMed]
- Stinchcombe TE. Current management of RET rearranged non-small cell lung cancer. Ther Adv Med Oncol 2020;12:1758835920928634. [Crossref] [PubMed]
- Lu C, Dong XR, Zhao J, et al. Association of genetic and immuno-characteristics with clinical outcomes in patients with RET-rearranged non-small cell lung cancer: a retrospective multicenter study. J Hematol Oncol 2020;13:37. [Crossref] [PubMed]
- Offin M, Guo R, Wu SL, et al. Immunophenotype and Response to Immunotherapy of RET-Rearranged Lung Cancers. JCO Precis Oncol 2019;3:PO.18.00386. [Crossref] [PubMed]
- Ferrara R, Auger N, Auclin E, et al. Clinical and Translational Implications of RET Rearrangements in Non-Small Cell Lung Cancer. J Thorac Oncol 2018;13:27-45. [Crossref] [PubMed]
- Baker JA, Sireci AN, Marella N, et al. Analytical Accuracy of RET Fusion Detection by Break-Apart Fluorescence In Situ Hybridization. Arch Pathol Lab Med 2022;146:351-9. [Crossref] [PubMed]
- Yang SR, Aypar U, Rosen EY, et al. A Performance Comparison of Commonly Used Assays to Detect RET Fusions. Clin Cancer Res 2021;27:1316-28. [Crossref] [PubMed]
- Xiang C, Guo L, Zhao R, et al. Identification and Validation of Noncanonical RET Fusions in Non-Small-Cell Lung Cancer through DNA and RNA Sequencing. J Mol Diagn 2022;24:374-85. [Crossref] [PubMed]
- Li W, Zhang Z, Guo L, et al. Assessment of cytology based molecular analysis to guide targeted therapy in advanced non-small-cell lung cancer. Oncotarget 2016;7:8332-40. [Crossref] [PubMed]
- Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228-47. [Crossref] [PubMed]
- Chu YH, Wirth LJ, Farahani AA, et al. Clinicopathologic features of kinase fusion-related thyroid carcinomas: an integrative analysis with molecular characterization. Mod Pathol 2020;33:2458-72. [Crossref] [PubMed]
- Skálová A, Ptáková N, Santana T, et al. NCOA4-RET and TRIM27-RET Are Characteristic Gene Fusions in Salivary Intraductal Carcinoma, Including Invasive and Metastatic Tumors: Is "Intraductal" Correct?. Am J Surg Pathol 2019;43:1303-13. [Crossref] [PubMed]
- Weisman PS, Altinok M, Carballo EV, et al. Uterine Cervical Sarcoma With a Novel RET-SPECC1L Fusion in an Adult: A Case Which Expands the Homology Between RET-rearranged and NTRK-rearranged Tumors. Am J Surg Pathol 2020;44:567-70. [Crossref] [PubMed]
- Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med 2012;18:378-81. [Crossref] [PubMed]
- Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med 2012;18:382-4. [Crossref] [PubMed]
- Pan Y, Zhang Y, Li Y, et al. ALK, ROS1 and RET fusions in 1139 lung adenocarcinomas: a comprehensive study of common and fusion pattern-specific clinicopathologic, histologic and cytologic features. Lung Cancer 2014;84:121-6. [Crossref] [PubMed]
- Saad AG, Yeap BY, Thunnissen FB, et al. Immunohistochemical markers associated with brain metastases in patients with nonsmall cell lung carcinoma. Cancer 2008;113:2129-38. [Crossref] [PubMed]
- Wang H, Wang Z, Zhang G, et al. Driver genes as predictive indicators of brain metastasis in patients with advanced NSCLC: EGFR, ALK, and RET gene mutations. Cancer Med 2020;9:487-95. [Crossref] [PubMed]
- Subbiah V, Velcheti V, Tuch BB, et al. Selective RET kinase inhibition for patients with RET-altered cancers. Ann Oncol 2018;29:1869-76. [Crossref] [PubMed]
- Drilon A, Oxnard GR, Tan DSW, et al. Efficacy of Selpercatinib in RET Fusion-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2020;383:813-24. [Crossref] [PubMed]
- Gainor JF, Curigliano G, Kim DW, et al. Pralsetinib for RET fusion-positive non-small-cell lung cancer (ARROW): a multi-cohort, open-label, phase 1/2 study. Lancet Oncol 2021;22:959-69. [Crossref] [PubMed]
- Takeuchi K. Discovery Stories of RET Fusions in Lung Cancer: A Mini-Review. Front Physiol 2019;10:216. [Crossref] [PubMed]
- Arai S, Kita K, Tanimoto A, et al. In vitro and in vivo anti-tumor activity of alectinib in tumor cells with NCOA4-RET. Oncotarget 2017;8:73766-73. [Crossref] [PubMed]
- Wang M, Naganna N, Sintim HO. Identification of nicotinamide aminonaphthyridine compounds as potent RET kinase inhibitors and antitumor activities against RET rearranged lung adenocarcinoma. Bioorg Chem 2019;90:103052. [Crossref] [PubMed]
- Lee MS, Kim RN. Identification of a novel partner gene, KIAA1217, fused to RET: Functional characterization and inhibitor sensitivity of two isoforms in lung adenocarcinoma. Oncotarget 2016;7:36101-14. [Crossref] [PubMed]
- Drilon A, Rekhtman N, Arcila M, et al. Cabozantinib in patients with advanced RET-rearranged non-small-cell lung cancer: an open-label, single-centre, phase 2, single-arm trial. Lancet Oncol 2016;17:1653-60. [Crossref] [PubMed]
- Lee SH, Lee JK, Ahn MJ, et al. Vandetanib in pretreated patients with advanced non-small cell lung cancer-harboring RET rearrangement: a phase II clinical trial. Ann Oncol 2017;28:292-7. [Crossref] [PubMed]
- Subbiah V, Yang D, Velcheti V, et al. State-of-the-Art Strategies for Targeting -Dependent Cancers. J Clin Oncol 2020;38:1209-21. [Crossref] [PubMed]
- Lin JJ, Zhu VW, Yoda S, et al. Impact of EML4-ALK Variant on Resistance Mechanisms and Clinical Outcomes in ALK-Positive Lung Cancer. J Clin Oncol 2018;36:1199-206. [Crossref] [PubMed]
- Canale M, Petracci E, Delmonte A, et al. Impact of TP53 Mutations on Outcome in EGFR-Mutated Patients Treated with First-Line Tyrosine Kinase Inhibitors. Clin Cancer Res 2017;23:2195-202. [Crossref] [PubMed]
- Canale M, Petracci E, Delmonte A, et al. Concomitant TP53 Mutation Confers Worse Prognosis in EGFR-Mutated Non-Small Cell Lung Cancer Patients Treated with TKIs. J Clin Med 2020;9:1047. [Crossref] [PubMed]
- Labbé C, Cabanero M, Korpanty GJ, et al. Prognostic and predictive effects of TP53 co-mutation in patients with EGFR-mutated non-small cell lung cancer (NSCLC). Lung Cancer 2017;111:23-9. [Crossref] [PubMed]
- Liu S, Yu J, Zhang H, et al. TP53 Co-Mutations in Advanced EGFR-Mutated Non-Small Cell Lung Cancer: Prognosis and Therapeutic Strategy for Cancer Therapy. Front Oncol 2022;12:860563. [Crossref] [PubMed]
- Wang WX, Xu CW, Chen YP, et al. TP53 mutations predict for poor survival in ALK rearrangement lung adenocarcinoma patients treated with crizotinib. J Thorac Dis 2018;10:2991-8. [Crossref] [PubMed]
- Costa DB. TP53 mutations are predictive and prognostic when co-occurring with ALK rearrangements in lung cancer. Ann Oncol 2018;29:2028-30. [Crossref] [PubMed]
- Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer 2009;9:701-13. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Sun Y, Pei L, Luo N, et al. A Novel MYH9-RET Fusion Occurrence and EGFR T790M Loss as an Acquired Resistance Mechanism to Osimertinib in a Patient with Lung Adenocarcinoma: A Case Report. Onco Targets Ther 2020;13:11177-81. [Crossref] [PubMed]
- Kim M, Na JM, Lee GW, et al. EGFR-mutated pulmonary adenocarcinoma with concurrent PIK3CA mutation, and with acquired RET fusion and EGFR T790M mutation after afatinib therapy. J Pathol Transl Med 2021;55:79-82. [Crossref] [PubMed]
- McCoach CE, Le AT, Gowan K, et al. Resistance Mechanisms to Targeted Therapies in ROS1+ and ALK+ Non-small Cell Lung Cancer. Clin Cancer Res 2018;24:3334-47. [Crossref] [PubMed]
- Drilon A, Bergagnini I, Delasos L, et al. Clinical outcomes with pemetrexed-based systemic therapies in RET-rearranged lung cancers. Ann Oncol 2016;27:1286-91. [Crossref] [PubMed]
- Tan AC, Seet AOL, Lai GGY, et al. Molecular Characterization and Clinical Outcomes in RET-Rearranged NSCLC. J Thorac Oncol 2020;15:1928-34. [Crossref] [PubMed]
- Shaw AT, Felip E, Bauer TM, et al. Lorlatinib in non-small-cell lung cancer with ALK or ROS1 rearrangement: an international, multicentre, open-label, single-arm first-in-man phase 1 trial. Lancet Oncol 2017;18:1590-9. [Crossref] [PubMed]
- Radonic T, Geurts-Giele WRR, Samsom KG, et al. RET Fluorescence In Situ Hybridization Analysis Is a Sensitive but Highly Unspecific Screening Method for RET Fusions in Lung Cancer. J Thorac Oncol 2021;16:798-806. [Crossref] [PubMed]
- Tsuta K, Kohno T, Yoshida A, et al. RET-rearranged non-small-cell lung carcinoma: a clinicopathological and molecular analysis. Br J Cancer 2014;110:1571-8. [Crossref] [PubMed]
- Belli C, Penault-Llorca F, Ladanyi M, et al. ESMO recommendations on the standard methods to detect RET fusions and mutations in daily practice and clinical research. Ann Oncol 2021;32:337-50. [Crossref] [PubMed]
- Mizukami T, Shiraishi K, Shimada Y, et al. Molecular mechanisms underlying oncogenic RET fusion in lung adenocarcinoma. J Thorac Oncol 2014;9:622-30. [Crossref] [PubMed]
- Li W, Guo L, Liu Y, et al. Potential Unreliability of Uncommon ALK, ROS1, and RET Genomic Breakpoints in Predicting the Efficacy of Targeted Therapy in NSCLC. J Thorac Oncol 2021;16:404-18. [Crossref] [PubMed]


