
Gene expression profiling of circulating tumor cells captured by MicroCavity Array is superior to enumeration in demonstrating therapy response in patients with newly diagnosed advanced and locally advanced non-small cell lung cancer
Introduction
Lung cancer is the second most common cancer in both males and females, with non-small cell lung cancer (NSCLC) being the most common (87% of cases) (1). It is by far the leading cause of cancer deaths among both men and women, making up almost 25% of all cancer deaths. Each year, more people die of lung cancer than of colon, breast, and prostate cancers combined (Cancer.org). The high mortality rate is often attributed to disease dissemination for which circulating tumor cells (CTCs) are the main precursors. CTCs are one of the mainstays of liquid biopsy (2) which also includes circulating tumor DNA (ctDNA), cell-free RNA, extracellular vesicles and tumor educated platelets. Liquid biopsy has more recently entered clinical practice through ctDNA as a companion diagnostic for targeted therapy of NSCLC (Guardant 360 CDx, Redwood City, CA, USA) and breast cancer, metastatic castrate resistant prostate cancer, ovarian cancer and NSCLC (FoundationOne Liquid CDx, Cambridge, MA, USA).
CTCs afford several advantages in providing greater insights into the disease state, especially considering the fact that they offer a broad sampling of both primary and metastatic sites. This is accomplished in a non-invasive blood collection during standard of care (SOC) throughout active treatment and monitoring period. Intratumor heterogeneity and tumor evolution results in resistance to therapy leading to poor outcomes, and tumor diversity is more truly reflected by diversity in CTCs. CTCs therefore could be much more effective clinical tools for prognostic stratification of patients. Since CTCs are live cells, they can be used in drug screens and functional assays. More importantly, they may predict relapse after primary treatment (3) even in early stage NSCLC (4).
CELLSEARCH (Menarini Silicon Biosystems, Bologna, Italy) is the only Food and Drug Administration (FDA) approved CTC technology to be implemented in clinical settings for breast, prostate and colorectal cancers. Unfortunately, CTC enumeration has demonstrated little value in clinical utility compared with SOC imaging technologies. The Southwest Oncology Group (SWOG) S0500 trial in metastatic breast cancer patients failed to demonstrate that an early change in chemotherapy based on the persistence of ≥5 CTCs per 7.5 mL of blood after one cycle of initial chemotherapy can improve overall survival (OS), as compared with a treatment change based on radiological detection of disease progression (5). While SWOG S0500 confirmed that CTC enumeration is an effective prognostic tool in determining progression free survival (PFS) and OS, it also suggested that CTC enumeration is ineffective as a solitary tool for treatment decisions. We and others have found that gene expression is a viable alternative to visual enumeration for detection of CTC (6-11). These studies highlight the need to incorporate molecular signatures of CTCs to complement enumeration from liquid biopsy. More recent studies also suggest that the heterogeneity of tumor cells traversing the blood stream may include cells that are not captured by the commonly used epithelial-targeted immunoaffinity enrichment of CTC. Hence, there is a need to enrich cells agnostic of cell surface proteins for an unbiased characterization for CTCs and the underlying disease state. In this study, we used the MicroCavity Array (MCA) (12-15), platform that captures CTCs from blood based on size without any preference for specific markers such as epithelial cell adhesion molecule (EPCAM) in NSCLC patients undergoing chemoradiation.
In this study, CTC enumeration had minimal prognostic value for patients with NSCLC undergoing chemoradiation. However, analysis of gene expression by enriched CTCs offered superior prognostic value prior to therapy and for monitoring disease. Also, we demonstrate that CTC gene expression patterns could make significant strides in non-invasive NSCLC management in the clinic. We present the following article in accordance with the Materials Design Analysis Reporting (MDAR) reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-314/rc).
Methods
Patients and healthy volunteers
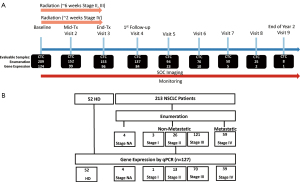
Patients with locally advanced or advanced NSCLC were recruited prior to initiation of radiation/chemo-radiation from the Thoracic Center at the University of Texas MD Anderson Cancer Center (Table 1). Patients were recruited under protocol Lab09-0307; healthy donor (HD) volunteers older than 18 years of age were recruited under protocol PA14-0063. All patients received chemoradiation as SOC treatment and all patients are not resected. Blood samples from NSCLC patients were collected prior to initiation of radiation (baseline), midway through radiation, at or soon after the conclusion of radiation, at the first follow-up after radiation and at 3-month intervals thereafter and as available through 24 months after enrollment. Patients enrolled with metastatic disease were typically treated with radiation in consolidation with shorter treatments of about 2 weeks. For these patients, samples collected before the end of radiation were coded as “midway through radiation” and samples collected at or after the completion of radiation, but before the 6-month follow-up were coded as “end of radiation therapy”. Radiation doses of 1,250 to 7,260 cGy were delivered via photons or protons. Self-reported cancer-free HD were used as controls (82 samples from 52 donors). A total of 213 patients were enrolled in the study between May 2016 and Dec 2020 contributing a total of 916 samples. All research blood samples were drawn along with routine clinical blood samples as per treatment schedule. Four patients missed baseline samples. Study outcomes did not affect the clinical management of the enrolled patients. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and its subsequent amendments. The study was approved by the Institutional Review Board of The University of Texas MD Anderson Cancer Center (protocols Lab09-0307 and PA14-0063) and informed consent was taken from all individual participants.
Table 1
| Characteristics | Subtypes | CTC enumeration (N=213) | CTC gene expression (N=127) |
|---|---|---|---|
| Age, years | Mean 67.0 (range, 37–88) | Mean 66.0 (range, 37–83) | |
| Gender | Female | 89 (41.8) | 56 (44.1) |
| Male | 124 (58.2) | 71 (55.9) | |
| ECOG | 0 | 59 (27.7) | 36 (28.3) |
| 1 | 130 (61.0) | 78 (61.4) | |
| 2 | 24 (11.3) | 13 (10.2) | |
| Smoker | No | 39 (18.3) | 24 (18.9) |
| Yes | 174 (81.7) | 103 (81.1) | |
| Histology | Unknown | 1 (0.5) | 0 (0.0) |
| NSCLC | 211 (99.1) | 126 (99.2) | |
| Neuro | 1 (0.5) | 1 (0.8) | |
| T | Unknown | 25 (11.7) | 14 (11.0) |
| T1 | 38 (17.8) | 20 (15.7) | |
| T2 | 57 (26.8) | 28 (22.0) | |
| T3 | 46 (21.6) | 34 (26.8) | |
| T4 | 37 (17.4) | 24 (18.9) | |
| Tx | 10 (4.7) | 7 (5.5) | |
| N | Unknown | 24 (11.3) | 13 (10.2) |
| N0 | 35 (16.4) | 22 (17.3) | |
| N1 | 27 (12.7) | 17 (13.4) | |
| N2 | 86 (40.4) | 51 (40.2) | |
| N3 | 33 (15.5) | 19 (15.0) | |
| Nx | 8 (3.8) | 5 (3.9) | |
| Stage | Unknown | 4 (1.9) | 4 (3.1) |
| I | 3 (1.4) | 1 (0.8) | |
| II | 26 (12.2) | 13 (10.2) | |
| III | 121 (56.8) | 70 (55.1) | |
| IV | 59 (27.7) | 39 (30.7) | |
| Induction chemo | No | 178 (83.6) | 104 (81.9) |
| Yes | 35 (16.4) | 23 (18.1) | |
| Radiation therapy modality | Photon | 164 (77.0) | 103 (81.1) |
| Proton | 49 (23.0) | 24 (18.9) |
CTC, circulating tumor cell; ECOG Eastern Cooperative Oncology Group Performance Status Scale; T, tumor size; N, nodal status; M, metastatic status; NSCLC, non-small cell lung cancer; Neuro, neuroendocrine carcinoma.
Sample processing
Peripheral blood samples were collected from patients and HDs into 10 mL ethylenediamine tetraacetic acid (EDTA) blood collection tubes at each time point. CTC enrichment was performed using an MCA designed and manufactured by Showa Denko Materials Co., Ltd. (formerly Hitachi Chemical, Tokyo, Japan) as described previously (16). Enrichment procedures for enumeration and molecular characterization were performed in parallel on separate aliquots of the same blood sample of about 9.5 mL each. The MCA closed chip was used for in situ staining and enumeration while the MCA open chip was used to enrich cells for subsequent gene expression analysis. For gene expression, whole blood was depleted of leukocytes with anti-CD45 beads (Miltenyi Biotech, Bergisch Gladbach, Germany) prior to enrichment of CTCs by MCA. In contrast to the prior report (16), cells enriched for gene expression analysis were not subjected to red blood cell (RBC) lysis for removal of contaminating RBCs as further optimization showed improved polymerase chain reaction (PCR) performance without RBC lysis. The resulting enriched cells were lysed in QIAzol and the lysates were archived at −80 °C. The first 86 patients enrolled through 11/2017 and 260 follow-up samples enrolled through 12/2020 were evaluated for CTC enumeration. The following 127 patients enrolled 11/2017 through 12/2020 were processed for both enumeration and gene expression from CTC-enriched lysates including 320 follow-up samples. Similar to the patient samples, each HD sample of 9.5 mL of blood was depleted of leukocytes and subsequently processed by MCA system.
Sample processing for enumeration of captured CTC
CTC-enriched fractions were stained in situ with a cocktail of antibodies to epithelial cytokeratins (CK) and leukocytes (CD45) and subjected to image analysis (16). CTCs in patient samples were counted in “real-time” at the time of imaging. Identification of CTCs was made based on guidelines published by Zeune and his co-workers (17). Generally, closed chips were imaged within 2 days of blood draw (mean turn-around-time was 1.6±1.8 days) using an Olympus IX81-DSU system (Olympus, Tokyo, Japan; The Flow Cytometry and Cellular Imaging Core Facility at MD Anderson NCI Cancer Center Support Grant P30CA16672). Captured 10× images, along with bright-field morphology, were used to identify CTCs, defined as nucleated cells (stained with 4’,6-diamidino-2-phenylindole, DAPI+) that were CK+ and CD45−.
Sample processing for gene expression of enriched CTC
Following white blood cell (WBC) depletion and automated enrichment of CTCs by the MCA open chip, samples were lysed with a phenol/guanidine thiocyanate solution (QIAzol, Qiagen, Venlo, Netherlands) and the lysate archived at −80 °C. Towards the conclusion of the study, HD and patient samples were processed for gene expression analysis in batch. Only samples of patients from the first 4 clinical visits were analyzed by quantitative real-time PCR (qRT-PCR). HD samples were randomized with the patient samples and tested concurrently to minimize inter-class variability. For RNA extraction, samples were processed in batches of 12 with HD samples randomized with the patient samples. RNA was isolated using phenol/chloroform extraction followed by Qiagen RNeasy Plus Micro kit automated on a QIAcube liquid handler for consistency. The cDNA was prepared by reverse-transcriptase and frozen at −20 °C. qRT-PCR was performed on 6 samples at a time using custom-designed BioRad PrimePCR plates that included internal controls for RNA quality, efficiency of reverse transcription, genomic DNA contamination, and a positive PCR control in every plate. The panel also housed controls for intra-plate variability and well-to-well liquid carry-over. Primers were designed following the principles established by the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines (18) and wet lab-validated by the manufacturer. Each gene was measured as a technical triplicate. PCR setup was performed using a QIAgility liquid handler (Qiagen).
Twenty target genes including 15 CTC-associated genes, 3 housekeeping control genes, and 2 hematopoietic control genes were evaluated by qRT-PCR to characterize cells of epithelial, mesenchymal, and cancer stem cell lineage. The panel included genes related to epithelial characteristics (CDH1, EGFR, EPCAM, KRT7, KRT18, MUC1), epithelial to mesenchymal transition (EMT) characteristics (AXL, FN1, SNAI2), cancer stem-like lineage (ALDH1A1), and signaling pathways commonly perturbed in NSCLC [BCL2, CD274/programmed death-ligand 1 (PD-L1), ERBB2, FGFR1, and MET]. Control genes included PTPRC (CD45) as a WBC control, GYPA as a nucleated RBC control, and housekeeping genes B2M, GAPDH and HPRT as positive controls (Table S1).
Statistical analysis
Blood samples were collected from patients with NSCLC prior to initiation of radiation/chemoradiation therapy (baseline), midway through radiation therapy, at the conclusion of radiation therapy, at the first follow-up after radiation therapy and at 3-month intervals thereafter, as available through 24 months after enrollment. Wilcoxon signed-rank test was used to evaluate the measurement changes at different time points. Mixed model was applied to assess the association between the repeated measurements (e.g., CTC) and clinical outcome after considering the within-patient correlation. OS is defined as the time from the start of radiation treatment till death or last follow-up. PFS is defined as the start of radiation treatment till relapse/death or last follow-up. It should be noted that there is no established gold-standard for CTC counts in NSCLC, although others have reported the presence of any CTCs, 3 CTCs, 5 CTC or 7 CTCs resulted in inferior PFS and OS (16,19-22). Rarefaction curve analysis was used to determine the optimal CTC count to be used as a cut off, but we were unable to find a satisfactory threshold and hence used a cut off of 5 CTC as established in metastatic breast and prostate cancers for further analysis. CTC count was evaluated as a continuous variable and dichotomized into <5 or ≥5 enumerated CTCs per sample (9.5 mL of blood). Gene expression using qRT-PCR was evaluated as a continuous variable of 40-Ct (cycle threshold), such that each 1-unit change represented a two-fold increase in expression and was not normalized. A similar analysis was performed for samples collected at the first follow-up visit following completion of radiation therapy. Kaplan-Meier curves were used to estimate the survival function for OS and PFS. Log rank tests were used to compare each time-to-event variable between groups. The analysis focuses primarily on the impact of the baseline (prior to initiation of chemoradiation therapy) CTC counts and CTC gene expression on survival. The Cox proportional hazards regression model was used to evaluate the corresponding effect on OS or PFS. For genes with baseline expression that strongly associated with survival, the time-dependent Cox regression for repeated measurement (e.g., gene expression) was applied to evaluate how the longitudinal measurements impact OS and PFS after adjusting other important clinical factors. The statistical analyses were carried out in SAS version 9.4 with figures and additional analyses generated in R version 3.6 (R Foundation for Statistical Computing, Vienna, Austria).
Results
A total of 213 NSCLC patients were enrolled contributing a total of 916 blood samples and 52 HD contributed a total of 82 blood samples. Patients presented with a range of disease stages as depicted in Table 1, including 154 patients with non-metastatic disease (stages I, II and III) and 59 with metastatic disease (stage IV) who received radiation as consolidation or stereotactic body radiation therapy (SBRT) with systemic therapy. During the follow-up through December 2020, 78 patients had died, and 105 patients had disease progression. A subset of samples from patients enrolled after November 2017 were interrogated for gene expression of the CTC-enriched cells including 127 patients and 320 longitudinal samples.
NSCLC patients primarily with stage II, III (non-metastatic) or stage IV (metastatic) disease were recruited prior to radiation/chemo-radiation treatment. Patients typically donated blood samples at baseline, immediately prior to initiation of radiation therapy, midway through radiation therapy, at the end of radiation therapy, at the first clinical follow-up, and at 3-month intervals as available thereafter (Figure 1A). For gene expression analysis, the samples at baseline through first follow-up were the primary focus (Figure 1B). Of the 154 patients with non-metastatic disease, 100 provided a first follow-up, post radiation (Visit 4) sample; of the 59 patients with metastatic disease, 39 provided a first follow-up, post radiation (Visit 4) sample. Patients recruited for this study included those that responded to induction chemotherapy and those that did not. Therefore, at study baseline, survival was independent of stage (Figure S1).
Primary endpoint of the clinical trial: CTC enumeration
Distribution of CTC counts
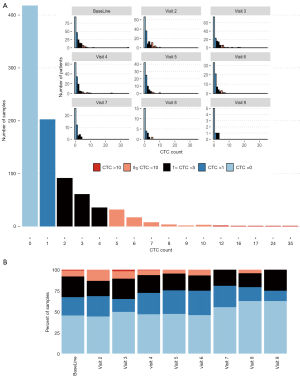
Identification of CTCs was made based on guidelines published by Zeune and his co-workers (17) as CK+ CD45− nucleated cells. As seen in Figure 2A,2B, CTC counts from image analyses suggest that at baseline, about 45% of blood samples from NSCLC patients have 0 CTCs and about 67% of all NSCLC samples have 0 or 1 CTC. Most patients have fewer than 5 CTCs (91%). There is a high level of patient attrition at later timepoints such that the statistical weight of each surviving individual increases at later timepoints and survivorship bias likely influences the distribution presented. There was no difference in CTC counts between patients with metastatic and non-metastatic disease. The percent of samples with observable CTCs is lowest in the samples collected at least 1 year after enrollment (Visit 7 and later) as the patients available for follow-up are likely the ones with good responses. The highest CTC count observed was 35 CTCs in ~9.5 mL of blood from a baseline sample. Compared to similarly enriched CTCs from patients with metastatic breast cancer, CTCs from NSLC patients exhibit less uniformity in CK staining and are relatively smaller and more challenging to identify (data not shown). As the data demonstrate, enumeration of CTCs from patients with NSCLC proved to be challenging as the morphology was poorly conducive to optical/visual recognition.
Survival by CTC enumeration
Comparison of patients who experienced disease progression with patients who did not have progression during the study showed minimal differences in CTC counts (e.g., baseline Student’s t-test P=0.85), although there was a marginally lower proportion of patients with 0 CTCs in patients with progression (Figure S2). Mixed model analysis employed to assess the association between longitudinal CTC enumeration and clinical outcome after considering the within-patient correlation showed that patients who experienced disease progression did not have significantly different CTC counts (Table 2). This analysis showed there was a slight decrease in counts over time irrespective of response to treatment.
Table 2
| Variables | CTC count change over time based on mixed model | P value | |
|---|---|---|---|
| Estimate | Standard error | ||
| Progression: progressed vs. non-progression | 0.259 | 0.201 | 0.1987 |
| Radiation treatment: Proton vs. Photon | 0.272 | 0.236 | 0.2489 |
| Time (months) | −0.045 | 0.012 | 0.0003 |
CTC, circulating tumor cell.
Rarefaction curve analysis observing PFS and OS at different CTC cutoffs showed no clear trend (not shown). Overall, CTC counts as a continuous or as dichotomized variable mostly failed to reflect clinical prognosis at baseline (Table 3 and Figure 3A). Although CTC enumeration showed minimal prognostic value prior to treatment, at the first follow-up following completion of therapy (Visit 4), the risk of death increased with increasing CTC counts (Table 3). The HR for each additional CTC observed at this time point was 1.11 [95% confidence interval (CI): 1.00–1.24, P=0.043]. This suggests that detection of minimal residual disease by CTC enumeration may be useful to evaluate response to chemoradiation therapy but may not predict response prior to therapy.
Table 3
| Timepoint | Outcome | CTC threshold | HR (95% CI) | P value |
|---|---|---|---|---|
| Baseline | PFS | 1-unit change | 0.99 (0.95, 1.05) | 0.984 |
| >0 | 1.21 (0.81, 1.8) | 0.343 | ||
| ≥5 | 0.86 (0.41, 1.78) | 0.688 | ||
| OS | 1-unit change | 1.01 (0.95, 1.06) | 0.811 | |
| >0 | 1.49 (0.92, 2.40) | 0.102 | ||
| ≥5 | 1.02 (0.47, 2.23) | 0.951 | ||
| Visit 4 | PFS | 1-unit change | 1.06 (0.95, 1.17) | 0.255 |
| >0 | 1.06 (0.64, 1.74) | 0.821 | ||
| ≥5 | 1.01 (0.40, 2.53) | 0.981 | ||
| OS | 1-unit change | 1.11 (1.00, 1.24) | 0.043* | |
| >0 | 1.60 (0.86, 3.02) | 0.139 | ||
| ≥5 | 1.60 (0.49, 5.18) | 0.436 |
*, P<0.05. PFS, progression-free survival; OS, overall survival; CTC, circulating tumor cell; HR, hazard ratio; CI, confidence interval.
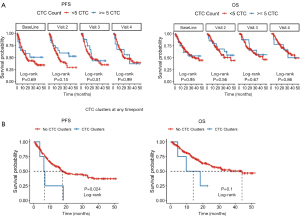
In our study, although imaging individual CTC in NSCLC yielded marginal, if any, clinical utility, detection of CTC clusters at any timepoint by imaging (Figure 3B) was highly correlated with poor prognosis, highlighting the benefit of enriching CTCs in NSCLC patients by size. We found CTC clusters in samples from 4 patients: 2 were found in baseline samples, one at the end of radiation treatment and one at the first follow-up following therapy. Detection of clusters at any of these timepoints was associated with an increased risk of progression (Cox proportional HR =3.00, 95% CI: 1.1–8.2, P=0.0318). Furthermore, as explained below, the 4 samples with clusters in our study were negative for EPCAM expression by qRT-PCR. Hence, the need to employ agnostic CTC enrichment for NSCLC is important for CTCs to be a prognostic tool.
In conclusion, although CTC enumeration did not provide meaningful clinical utility in this cohort of NSCLC patients at baseline, higher CTC counts after radiation therapy were associated with increased mortality.
Secondary endpoint of the clinical trial:gene expression of enriched CTCs
As a complementary/alternative measure of CTC burden, samples from a subset of patients enrolled in the study were subjected to targeted gene expression analysis. In this portion of the study, 124 NSCLC patients had baseline samples available for analysis and 84 at the first follow-up visit following completion of radiation (Visit 4).
Description of distribution
Gene expression overview is shown in Figure 4A. There was no significant difference in any gene expression between patients with metastatic disease and those with localized disease at baseline. Gene expression by CTC-enriched cells did not correlate with CTC enumeration (sum of gene expression Spearman rho =0.013, P=0.787) suggesting that gene expression may be an alternative, orthogonal measure to enumeration. Interestingly, the mesenchymal gene SNAI2 had a very weak but significant negative correlation with CTC count (Spearman rho =−0.106, P=0.0361). This was the only significant correlation of gene expression with enumerated count, suggesting that enriching CTC by size is able to pick up mesenchymal cells.

Gene expression was compared at each timepoint. During radiotherapy treatment (Visits 2 and/or 3) there was a slight but significant decrease in the mean expression of AXL, BCL2, ERBB2, KRT18 and MET compared to mean baseline expression (Figure 4B). For AXL, BCL2, and MET these drops are only in the patients who did not progress. Conversely, the decreases in ERBB2 were primarily in patients who progressed (Figure 4C).
For the full cohort at the first follow-up, none of the genes showed significantly lower mean expression compared to baseline (not shown), however there was a significant increase in mean CDH1 expression (not shown). We also looked at baseline-standardized expression which similarly lacked statistically significant variation in gene expression. However, patients who later progressed had significantly higher expression of EPCAM, ERBB2, KRT7, KRT18, and MUC1 (Wilcoxon signed-rank P<0.05) at both baseline and midway through radiation (Figure 4D). At the first follow-up after completion of radiation therapy, EPCAM, KRT7 and KRT18 remained elevated in patients who progressed (Figure 4D). These observations suggest that detection of CTC by gene expression is a viable alternative to visual enumeration.
To establish cut-offs for positive expression, HD blood processed through the MCA platform was used to define positive gene expression in the enriched samples. A cutoff for positive expression was established at 1.0 standard deviation above the mean expression for each gene in HD blood.
Univariate survival analysis
In contrast to CTC enumeration, detection of CTCs by gene expression was a strong and significant prognostic indicator of survival in this study. As a first pass, to select the most clinically interesting genes from the panel of 20 genes, all timepoints were combined in univariate Cox analysis for each gene without multiple comparison correction. In this analysis, expression of BCL2, CDH1, CD274, EPCAM, FGFR1, KRT18 and MUC1 was associated with an increased risk of progression and death with a trend in MET and FN1 (Figure S3). The associations of these nine genes with outcomes at individual timepoints are shown in Figure 5.
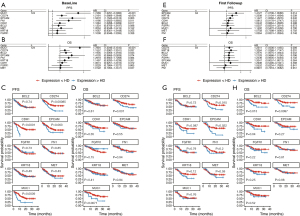
At baseline, high levels of CDH1, CD274 (PD-L1), EPCAM, FN1, and MUC1 were associated with an increased risk of progression (Figure 5A,5C) and BCL2, CD274, MUC1, were associated with an increased risk of death (Figure 5B,5D).
At the first clinical follow-up visit after completion of radiation therapy (Visit 4), when NSCLC patients could be evaluated for response to radiation therapy, expression of EPCAM, KRT18 and CD274 (PD-L1) were each associated with an increased risk of progression (Figure 5E,5G) and expression of BCL2, CD274 and KRT18 was associated with increased risk of death (Figure 5F,5H). Kaplan Meier curves at each timepoint are shown in Figure S4.
Alternatively, the number of positive genes can be considered as an attempt to capture the multiple heterogenous populations epithelial and mesenchymal CTC in a single number. Among the 9 genes with significant prognostic value, the greatest hazard of both progression and death was with the highest number of expressed genes in the CTC-enriched cells. This is summarized as the blue line in Figure 6A. Considering expressions of the 9 genes that were individually significant predictors of survival, increasing numbers of positive genes (x-axis) is associated with a decrease in PFS (red line in Figure 6A). With more than 4 positive genes, the hazard ratio (HR) climbs steeply (Figure 6A) although the number of patient samples with more than 4 positive genes drops significantly, thereby decreasing statistical power. Combined, this suggests gene expression is a more stable measure than CTC enumeration in these patients.
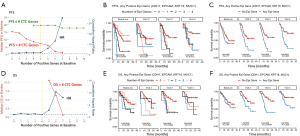
Looking at only the 4 primary epithelial genes (CDH1, EPCAM, KRT18, MUC1) there is a greater risk of progression with increasing number of positive genes detected in the CTC-enriched cells (Figure 6B). Patients with CTCs positive for at least 3 epithelial genes at baseline all progressed within 10 months (HR 8.2, P<0.001, 95% CI: 3.2–21.1); whereas those with 0 or 1 positive epithelial gene did not reach median survival. However, after initiation of therapy, there is minimal difference in survival among patients with varying numbers of positive genes (Figure 6B). In assessing response to therapy, if there is any positive epithelial gene in the CTC, the hazard of progression is much greater than if none are detected, and it does not matter if there are multiple genes detected (Figure 6C).
The number of positive epithelial genes detected is less impactful on OS and stratification by number is not significant at any of the first four timepoints (Figure 6D,6E). Nonetheless, using the higher sensitivity threshold of detection of any epithelial gene is useful in evaluating response to therapy. There is also an increased risk of death if any epithelial gene is detected midway through radiation (HR 2.2, P=0.0342), at the end of radiation (HR 3.7, P=0.00109) or at the first follow-up visit (HR 2.7, P=0.0323) (Figure 6F).
We have previously shown that PD-L1 can be tracked in CTC (23). Adding PD-L1 to the 4 epithelial genes, if any of the 5 are positive, the baseline measure has a significant negative prognosis (for risk of progression and death, Figure S5) whereas the 4 epithelial genes do not (Figure 6D,6F). However, after initiation of therapy, using the 5 genes (4 epithelial genes with PD-L1, Figure S5) is less prognostic than epithelial genes alone (Figure 6). Figure S5 shows the stratified risk of progression (Figure S5A) and death (Figure S5B) for the 5 genes and the risk if any of the 5 genes are positive (Figure S5C,S5D).
Whereas baseline detection of any epithelial gene offers minimal prognostic value (PFS HR 1.5, P=0.173, OS HR 1.3, P=0.387), effectively increasing the specificity by requiring at least two positive epithelial genes greatly increases the baseline HR (PFS HR for at least two genes at baseline 3.685, P<0.001, OS HR 2.4, P=0.0175). However, this increased specificity at baseline results in lower sensitivity that negates the prognostic value once on-treatment (Figure S6).
Multivariate survival analysis
BCL2, CD274 (PD-L1), EPCAM and MUC1 remained significant independent prognostic factors in multivariate, time-dependent analyses of OS (Table S2) and PFS (Table S3) that included Eastern Cooperative Oncology Group (ECOG) status, smoking status and stage where each gene was included in a separate model. Stage was removed from each of the final models as a non-significant factor.
Overall, this suggests that expression of CTC-related genes by enriched CTC is highly associated with clinical outcome.
Discussion
We demonstrated that CTC agnostically enriched by the MCA platform are prognostic in longitudinal analysis. However, unbiased gene-expression was better able to characterize the cells than enumeration by microscopy. Enumeration of CTCs from peripheral blood by image analysis following enrichment has been an accepted clinical practice for the past 15 years. Using the FDA-approved CELLSEARCH platform (developed by Veridex and currently marketed by Menarini Silicon Biosystems, Bologna, Italy), patients with metastatic breast cancer or metastatic prostate cancer with ≥5 CTCs per 7.5 mL of blood or patients with metastatic colorectal cancer with ≥3 CTCs per 7.5 mL of blood have been shown to have an unfavorable prognosis. To date, no cutoff for CTC count of prognostic or predictive value has been definitively established for NSCLC, although others have reported the presence of any CTCs, 3 CTCs, 5 CTC or 7 CTCs resulted in inferior PFS and OS (16,19-22). We were also unable to identify an optimal cutoff for this study and used a cutoff of 5 CTC for survival analysis. Overall, CTC counts as a continuous or as dichotomized variable mostly failed to reflect clinical prognosis at baseline. Other studies have found a lack of prognostic utility of CTCs in NSCLC (24-27).
Although single CTC had minimal clinical utility, detection of clusters at any timepoint was associated with an increased risk of progression. Similarly, the literature shows CTC cluster was significantly correlated with disease control rate of NSCLC (28). Also, Zeinali et al. showed that the majority of recovered CTCs/clusters from NSCLC patients were EPCAM-negative, suggesting that these CTCs would have been missed using traditional antibody-based capture methods (29). Hanssen et al. showed that EPCAM expression is low and less frequently detected in CTCs of NSCLC patients compared to other epithelial tumors (30,31). The 4 samples with clusters in our study were negative for EPCAM expression by qPCR. This highlights the need to employ agnostic enrichment for CTCs in NSCLC. As CTC clusters have been strongly attributed for the increased metastatic potential leading to poor prognosis in lung cancer (32) and other cancers (33-35) further study is warranted.
The widely used CELLSEARCH system enriches CTCs that express an EPCAM. However, EPCAM is frequently lost during metastatic progression in a process known as EMT. Therefore, many of the cells most responsible for the initiation of metastasis may be missed. In contrast, the MCA platform used in this study enriches CTCs based on size and deformability without a bias towards epithelial characteristics, potentially increasing the yield of clinically relevant CTCs. Indeed, gene expression by qPCR showed that the system enriched cells with mesenchymal characteristics.
Enumeration of CTCs by image analysis has been the gold standard liquid biopsy tool for monitoring patients with solid tumors of epithelial origin. However, counts are highly dependent on the operator’s analysis of the enriched cells. Moreover, since 2004 there has been an unmet clinical need for further molecular characterization beyond enumeration. As proof of concept that the MCA platform can enrich CTCs for molecular characterization, gene expression was included in a subset of patients in this study.
We found that gene expression by enriched CTC was a better prognostic tool than CTC enumeration. We noted that expression of epithelial genes was correlated with response to therapy. SNAI2 was the only gene with significant correlation of gene expression with enumerated count, suggesting that enriching CTC by size is able to pick up mesenchymal cells. Others have shown that this is important as CTCs with a “circulatory transition phenotype” are better able to survive the stress of circulation leading to increased metastatic spread and decreased survival (36). The expression of several genes such as BCL2, AXL, and MET decreased only in the patients who did not progress suggesting these signaling pathways were effectively targeted during therapy. The BCL2 correlation matches our previous observation of CTC in NSCLC (16). Others have seen that BCL2 (37), EPCAM (38) MUC1 (38) and PD-L1 (39) in CTCs are significant prognostic markers in NSCLC. As this study showed that molecular analysis of CTC is feasible, further development of DNA mutation analysis is on-going.
This study included a heterogeneous population of patients including patients with stage II, III and IV disease. The heterogeneity across patients helps us understand the relationship of CTC and prognosis or the molecular characteristics of CTC on prognosis across stages in patients who receive chemoradiation. All patients are not resected. In patients who received consolidation radiation, these patients have had prior chemotherapy exposure, so the CTCs are representative of a postinduction disease state. Although others have seen an association of CTC gene signatures with presence of metastasis (40) we did not observe any association with stage. This may be attributed to the patient cohort specific to this study, wherein patients with metastatic disease were recruited for radio therapy (and inclusion in this protocol) only after sufficient response to systemic therapy. Therefore, patients enrolled with higher stage disease may have had a better prognosis at study baseline.
SOC for NSCLC changed during the course of this study as immunotherapy was approved as an integral part of the treatment portfolio (41,42). Indeed, many patients included in this study received immunotherapy independent of radiotherapy. We showed that PD-L1 gene expression by CTC can be used to stratify patient risk prior to radiation. However, this study was not powered to interrogate the utility of CD274 expression during immunotherapy as PD-L1/PD-1 targeting agents were not initiated at baseline in 97% of these patients. Other studies have shown that PD-L1+ CTCs are associated with poor prognosis in NSCLC (39,43-45) and that PD-L1 expression increased at progression (46). However, in patients receiving immunotherapy, PD-L1 expression by CTC predicted better response to therapy (47,48). Furthermore, we have previously seen that CTCs are associated with abnormalities in peripheral blood immune cells (49,50) suggesting that further study is warranted.
Conclusions
We have been able to successfully detect CTCs at any timepoint in 83.6% of NSCLC patients enrolled in this study. Although we could not demonstrate the prognostic element of individual CTC enumeration in these patients, CTC clusters had significant prognostic potential. We could detect elevated CTC-associated genes in the baseline samples of 105 patients (84.7%) and could detect at least 1 elevated epithelial gene in 46.7% of baseline samples tested. Following patients across timepoints, 124 (97.6%) of them had at least 1 gene detectable in at least 1 sample, and 101 (79.5%) had at least one elevated epithelial gene in at least one timepoint. High expression of the following genes were markers of poor prognosis: BCL2, CD274 (PD-L1), CDH1, EPCAM, FGFR1, FN1, KRT18, MET and MUC1. Although enumeration of CTC by imaging is not an ideal clinical tool due to their morphology, data shows that CTC clusters and gene expression are powerful prognostic tools.
Acknowledgments
We wish to acknowledge Sanda Tin and James Aguilar for obtaining healthy donor blood and processing samples, respectively and Jianzhong He and Yawei Qiao for sample logistics. We would also like to thank Jared Burks for microscopy support.
Funding: This work was supported in part by The University of Texas MD Anderson’s Cancer Center Support Grant (P30CA016672), the National Cancer Institute’s (NCI) Research Specialist 1 R50 CA243707-01A1, and Showa Denko Materials Co., Ltd. (Previously Hitachi Chemical Co.) Japan.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-314/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-314/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-314/coif). SHL serves as an unpaid editorial board member of Translational Lung Cancer Research from October 2021 to September 2023. All authors report that Hitachi Chemical Company of Tokyo Japan (currently Showa Denko Materials Co., Ltd.) and The University of Texas MD Anderson Cancer Center entered into a Strategic Alliance Agreement to develop the MCA instrument and conduct the clinical trial that produced data reported in this manuscript. JMR serves as a member of the Scientific Advisory Board for Angle plc. SHL reports grants from STCube Pharmaceuticals, Beyond Spring Pharmaceuticals, Nektar Therapeutics, other support from Creatv Microtech, AstraZeneca Inc., XRAD Therapeutics, non-financial support from Scenexo, Inc., outside the submitted work. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and its subsequent amendments. The study was approved by the Institutional Review Board of The University of Texas MD Anderson Cancer Center (protocols Lab09-0307 and PA14-0063) and informed consent was taken from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Facts & Figures 2022. In: American Cancer Society. Bethesda, MD. 2022. Available online: https://seer.cancer.gov/statfacts/html/lungb.html. Accessed 4/21/2022.
- Christopoulos P. Liquid biopsies come of age in lung cancer. Transl Lung Cancer Res 2022;11:706-10. [Crossref] [PubMed]
- Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic - implementation issues and future challenges. Nat Rev Clin Oncol 2021;18:297-312. [Crossref] [PubMed]
- Shi J, Li F, Yang F, et al. The combination of computed tomography features and circulating tumor cells increases the surgical prediction of visceral pleural invasion in clinical T1N0M0 lung adenocarcinoma. Transl Lung Cancer Res 2021;10:4266-80. [Crossref] [PubMed]
- Smerage JB, Barlow WE, Hortobagyi GN, et al. Circulating tumor cells and response to chemotherapy in metastatic breast cancer: SWOG S0500. J Clin Oncol 2014;32:3483-9. [Crossref] [PubMed]
- Cohen EN, Jayachandran G, Hardy MR, et al. Antigen-agnostic microfluidics-based circulating tumor cell enrichment and downstream molecular characterization. PLoS One 2020;15:e0241123. [Crossref] [PubMed]
- Mego M, Cholujova D, Minarik G, et al. CXCR4-SDF-1 interaction potentially mediates trafficking of circulating tumor cells in primary breast cancer. BMC Cancer 2016;16:127. [Crossref] [PubMed]
- Mego M, Karaba M, Minarik G, et al. Circulating Tumor Cells With Epithelial-to-mesenchymal Transition Phenotypes Associated With Inferior Outcomes in Primary Breast Cancer. Anticancer Res 2019;39:1829-37. [Crossref] [PubMed]
- Andreopoulou E, Yang LY, Rangel KM, et al. Comparison of assay methods for detection of circulating tumor cells in metastatic breast cancer: AdnaGen AdnaTest BreastCancer Select/Detect versus Veridex CellSearch system. Int J Cancer 2012;130:1590-7. [Crossref] [PubMed]
- Mego M, Gao H, Lee BN, et al. Prognostic Value of EMT-Circulating Tumor Cells in Metastatic Breast Cancer Patients Undergoing High-Dose Chemotherapy with Autologous Hematopoietic Stem Cell Transplantation. J Cancer 2012;3:369-80. [Crossref] [PubMed]
- Mego M, Mani SA, Lee BN, et al. Expression of epithelial-mesenchymal transition-inducing transcription factors in primary breast cancer: The effect of neoadjuvant therapy. Int J Cancer 2012;130:808-16. [Crossref] [PubMed]
- Hosokawa M, Kenmotsu H, Koh Y, et al. Size-based isolation of circulating tumor cells in lung cancer patients using a microcavity array system. PLoS One 2013;8:e67466. [Crossref] [PubMed]
- Negishi R, Hosokawa M, Nakamura S, et al. Development of the automated circulating tumor cell recovery system with microcavity array. Biosens Bioelectron 2015;67:438-42. [Crossref] [PubMed]
- Yoshino T, Tanaka T, Nakamura S, et al. Evaluation of cancer cell deformability by microcavity array. Anal Biochem 2017;520:16-21. [Crossref] [PubMed]
- Yagi S, Koh Y, Akamatsu H, et al. Development of an automated size-based filtration system for isolation of circulating tumor cells in lung cancer patients. PLoS One 2017;12:e0179744. [Crossref] [PubMed]
- Cohen EN, Jayachandran G, Gao H, et al. Enumeration and molecular characterization of circulating tumor cells enriched by microcavity array from stage III non-small cell lung cancer patients. Transl Lung Cancer Res 2020;9:1974-85. [Crossref] [PubMed]
- Zeune LL, de Wit S, Berghuis AMS, et al. How to Agree on a CTC: Evaluating the Consensus in Circulating Tumor Cell Scoring. Cytometry A 2018;93:1202-6. [Crossref] [PubMed]
- Bustin SA, Beaulieu JF, Huggett J, et al. MIQE precis: Practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC Mol Biol 2010;11:74. [Crossref] [PubMed]
- Hofman V, Ilie MI, Long E, et al. Detection of circulating tumor cells as a prognostic factor in patients undergoing radical surgery for non-small-cell lung carcinoma: comparison of the efficacy of the CellSearch Assay and the isolation by size of epithelial tumor cell method. Int J Cancer 2011;129:1651-60. [Crossref] [PubMed]
- Juan O, Vidal J, Gisbert R, et al. Prognostic significance of circulating tumor cells in advanced non-small cell lung cancer patients treated with docetaxel and gemcitabine. Clin Transl Oncol 2014;16:637-43. [Crossref] [PubMed]
- Muinelo-Romay L, Vieito M, Abalo A, et al. Evaluation of Circulating Tumor Cells and Related Events as Prognostic Factors and Surrogate Biomarkers in Advanced NSCLC Patients Receiving First-Line Systemic Treatment. Cancers (Basel) 2014;6:153-65. [Crossref] [PubMed]
- Xu YH, Zhou J, Pan XF. Detecting circulating tumor cells in patients with advanced non-small cell lung cancer. Genet Mol Res 2015;14:10352-8. [Crossref] [PubMed]
- Adams DL, Adams DK, He J, et al. Sequential Tracking of PD-L1 Expression and RAD50 Induction in Circulating Tumor and Stromal Cells of Lung Cancer Patients Undergoing Radiotherapy. Clin Cancer Res 2017;23:5948-58. [Crossref] [PubMed]
- Carlsson A, Nair VS, Luttgen MS, et al. Circulating tumor microemboli diagnostics for patients with non-small-cell lung cancer. J Thorac Oncol 2014;9:1111-9. [Crossref] [PubMed]
- Mascalchi M, Falchini M, Maddau C, et al. Prevalence and number of circulating tumour cells and microemboli at diagnosis of advanced NSCLC. J Cancer Res Clin Oncol 2016;142:195-200. [Crossref] [PubMed]
- Wendel M, Bazhenova L, Boshuizen R, et al. Fluid biopsy for circulating tumor cell identification in patients with early-and late-stage non-small cell lung cancer: a glimpse into lung cancer biology. Phys Biol 2012;9:016005. [Crossref] [PubMed]
- Gallo M, De Luca A, Maiello MR, et al. Clinical utility of circulating tumor cells in patients with non-small-cell lung cancer. Transl Lung Cancer Res 2017;6:486-98. [Crossref] [PubMed]
- Li J. Significance of Circulating Tumor Cells in Nonsmall-Cell Lung Cancer Patients: Prognosis, Chemotherapy Efficacy, and Survival. J Healthc Eng 2021;2021:2680526. [Crossref] [PubMed]
- Zeinali M, Lee M, Nadhan A, et al. High-Throughput Label-Free Isolation of Heterogeneous Circulating Tumor Cells and CTC Clusters from Non-Small-Cell Lung Cancer Patients. Cancers (Basel) 2020;12:127. [Crossref] [PubMed]
- Hanssen A, Wagner J, Gorges TM, et al. Characterization of different CTC subpopulations in non-small cell lung cancer. Sci Rep 2016;6:28010. [Crossref] [PubMed]
- Hanssen A, Loges S, Pantel K, et al. Detection of Circulating Tumor Cells in Non-Small Cell Lung Cancer. Front Oncol 2015;5:207. [Crossref] [PubMed]
- Sawabata N, Susaki Y, Nakamura T, et al. Cluster circulating tumor cells in surgical cases of lung cancer. Gen Thorac Cardiovasc Surg 2020;68:975-83. [Crossref] [PubMed]
- Davis AA, Zhang Q, Gerratana L, et al. Association of a novel circulating tumor DNA next-generating sequencing platform with circulating tumor cells (CTCs) and CTC clusters in metastatic breast cancer. Breast Cancer Res 2019;21:137. [Crossref] [PubMed]
- Giuliano M, Shaikh A, Lo HC, et al. Perspective on Circulating Tumor Cell Clusters: Why It Takes a Village to Metastasize. Cancer Res 2018;78:845-52. [Crossref] [PubMed]
- Donato C, Szczerba BM, Scheidmann MC, et al. Micromanipulation of Circulating Tumor Cells for Downstream Molecular Analysis and Metastatic Potential Assessment. J Vis Exp 2019; [Crossref] [PubMed]
- Alvarado-Estrada K, Marenco-Hillembrand L, Maharjan S, et al. Circulatory shear stress induces molecular changes and side population enrichment in primary tumor-derived lung cancer cells with higher metastatic potential. Sci Rep 2021;11:2800. [Crossref] [PubMed]
- Messaritakis I, Nikolaou M, Politaki E, et al. Bcl-2 expression in circulating tumor cells (CTCs) of patients with small cell lung cancer (SCLC) receiving front-line treatment. Lung Cancer 2018;124:270-8. [Crossref] [PubMed]
- Zhu WF, Li J, Yu LC, et al. Prognostic value of EpCAM/MUC1 mRNA-positive cells in non-small cell lung cancer patients. Tumour Biol 2014;35:1211-9. [Crossref] [PubMed]
- Sinoquet L, Jacot W, Gauthier L, et al. Programmed Cell Death Ligand 1-Expressing Circulating Tumor Cells: A New Prognostic Biomarker in Non-Small Cell Lung Cancer. Clin Chem 2021;67:1503-12. [Crossref] [PubMed]
- Josefsson A, Larsson K, Månsson M, et al. Circulating tumor cells mirror bone metastatic phenotype in prostate cancer. Oncotarget 2018;9:29403-13. [Crossref] [PubMed]
- Antonia SJ, Villegas A, Daniel D, et al. Overall Survival with Durvalumab after Chemoradiotherapy in Stage III NSCLC. N Engl J Med 2018;379:2342-50. [Crossref] [PubMed]
- Manapov F, Kenndoff S, Käsmann L. NICOLAS, DETERRED and KEYNOTE 799: focus on escalation of conventionally fractionated chemoradiotherapy by immune checkpoint inhibition in unresectable stage III non-small cell lung cancer. Transl Lung Cancer Res 2022;11:702-5. [Crossref] [PubMed]
- Dong J, Zhu D, Tang X, et al. Detection of Circulating Tumor Cell Molecular Subtype in Pulmonary Vein Predicting Prognosis of Stage I-III Non-small Cell Lung Cancer Patients. Front Oncol 2019;9:1139. [Crossref] [PubMed]
- Acheampong E, Abed A, Morici M, et al. Evaluation of PD-L1 expression on circulating tumour cells in small-cell lung cancer. Transl Lung Cancer Res 2022;11:440-51. [Crossref] [PubMed]
- Manjunath Y, Upparahalli SV, Avella DM, et al. PD-L1 Expression with Epithelial Mesenchymal Transition of Circulating Tumor Cells Is Associated with Poor Survival in Curatively Resected Non-Small Cell Lung Cancer. Cancers (Basel) 2019;11:806. [Crossref] [PubMed]
- Ntzifa A, Strati A, Kallergi G, et al. Gene expression in circulating tumor cells reveals a dynamic role of EMT and PD-L1 during osimertinib treatment in NSCLC patients. Sci Rep 2021;11:2313. [Crossref] [PubMed]
- Ouyang Y, Liu W, Zhang N, et al. Prognostic significance of programmed cell death-ligand 1 expression on circulating tumor cells in various cancers: A systematic review and meta-analysis. Cancer Med 2021;10:7021-39. [Crossref] [PubMed]
- Dall’Olio FG, Gelsomino F, Conci N, et al. PD-L1 Expression in Circulating Tumor Cells as a Promising Prognostic Biomarker in Advanced Non-small-cell Lung Cancer Treated with Immune Checkpoint Inhibitors. Clin Lung Cancer 2021;22:423-31. [Crossref] [PubMed]
- Mego M, Gao H, Cohen EN, et al. Circulating tumor cells (CTCs) are associated with abnormalities in peripheral blood dendritic cells in patients with inflammatory breast cancer. Oncotarget 2017;8:35656-68. [Crossref] [PubMed]
- Mego M, Gao H, Cohen EN, et al. Circulating Tumor Cells (CTC) Are Associated with Defects in Adaptive Immunity in Patients with Inflammatory Breast Cancer. J Cancer 2016;7:1095-104. [Crossref] [PubMed]


