Transcriptional deregulation underlying the pathogenesis of small cell lung cancer
Introduction
Small cell lung cancer (SCLC) is one of the few cancers for which mutations in transcription regulators, namely RB and P53, are a primary genetic cause. Genomic analyses of patient SCLC tumors revealed that a majority of recurrent somatic alterations affect transcription factors and chromatin modifiers including members of MYC family, SOX2, MLL1/2, CREBBP-EP300, RBL2, and P73. Frequent alterations in these proteins indicate that transcriptional deregulation beyond the loss of RB and P53 underlies cellular transformation and malignant progression of SCLC, causing aberrant expression of a broad range of genes related to cell proliferation and growth signaling pathways. As the concept of targeting chromatic modifiers including BRD4 using small molecule inhibitors have recently emerged for SCLC, characterization of the deregulated transcription programs is critical for discovery of potential targets and defining mechanism of targeted therapy. Recent integrated approaches using functional genomics and genetics and advanced mouse models have enabled interrogation of aberrant transcription program during SCLC development, identifying key oncogenic drivers and their mechanisms of action and conducting preclinical studies involving targeting transcriptional alterations. This review summarizes recent progresses in our understanding of transcriptional deregulation in SCLC development and tumor heterogeneity.
Transcription deregulation for tumor initiation and early-stage progression
The most common alterations in the SCLC are chromosomal deletions, truncations and missense mutations in the genes encoding TP53 and RB, which are observed in up to 90% of tumors (1-4). This high prevalence of loss-of-function alterations supported the hypothesis that these genes were important tumor suppressors in SCLC development. This hypothesis was formally tested in the studies of genetically engineered mouse models (GEMMs) of SCLC in which deleting the two tumor suppressors resulted in lung tumors resembling SCLC with 100% penetrance. Below is a summary of what we have learned about the roles of these tumor suppressors in SCLC and what remains to be discovered.
Loss of RB and TP53 functions—the rate-limiting step for SCLC development
RB and TP53 are bona fide tumor suppressors whose inactivation is directly linked to tumorigenesis (5-10). RB is best known for its role in controlling cell cycle progression (5,11,12). RB directly binds to E2F family of transcription factors and recruits HDACs and other transcriptional repressor complex proteins such that it inhibits expression of a number of cell cycle-related proteins, including cyclins (5). Inactivation of RB is also associated with an increase in cell plasticity through failed regulation of cell proliferation and apoptotic signaling (5,13,14). TP53 plays an important role in maintaining genomic stability against genotoxic stresses, including DNA damage (15,16). P53 as a transcription factor positively regulates expression of a number of genes involved in cell cycle arrest, senescence, apoptosis, and DNA repair, that collectively promote genomic integrity and tumor suppression (17). As a result of the loss-of-function mutations in P53 affected cells accumulate potentially oncogenic mutations evade oncogene-induced senescence and cell death, and propagate aberrantly due to cell cycle deregulation (16,17). The near-universal alteration of RB and TP53 genes indicates that removal of their broad spectrum of tumor suppressor activity is essential for the genesis of SCLC. This idea was firmly validated by the study of an SCLC GEMM—Rb/p53-conditional mutant mice (18). Complete loss of Rb and p53 via Cre-mediated recombination of floxed alleles in the lung epithelium results in development of mouse SCLC, whereas incomplete deletion of either Rb or p53 allele produces only lung adenocarcinoma (18,19). The requirement of RB loss is seen in several neuroendocrine (NE) tumors, including retinoblastoma and pituitary tumor, suggesting that the tumor suppressive function of RB is in part related to regulating NE differentiation. In support of this idea, lung epithelium-specific Rb-knockout alone resulted in hyperplasia of NE cells (20).
The GEMM study supports the concept that the inactivation of RB and TP53 is the rate-limiting step in SCLC development. However, the long tumor latency (9–12 months after Cre-mediated deletion of Rb and p53) seen in the GEMM also implies a dependency on additional oncogenic events driving Rb/p53-mutant cells toward malignancy (Figure 1A) (18,19). Indeed, the Rb/p53-mutant cells isolated from the lung epithelium of GEMM one month after Cre infection did not transform spontaneously, although they did continue to proliferate in culture (21). These benign characteristics of early-stage mutant cells, termed precancerous cells of SCLC (preSC for short), are largely attributed to the lack of oncogenic events typically seen in SCLC such as L-MYC overexpression (Figure 1B). These findings suggest that the loss of RB and P53 functions primarily confers upon cells unlimited replicative potential and the capacity to evade cell senescence and death and that later other oncogenic alterations drive malignant progression. This idea has been central to recent efforts aiming to determine these oncogenic alterations, some of which are discussed below with emphasis on transcription regulators. Defining transcription networks that drive the tumorigenic progression of cells lacking RB and p53 will provide critical insight to strategies for tumor intervention, as the concept of restoring RB and P53 function in lung cancer has yet to be translated into clinical strategy (22-24).
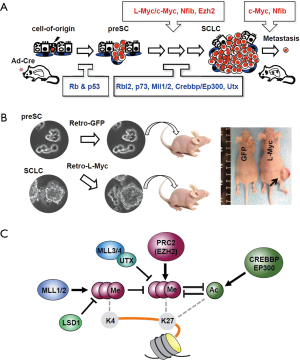
RBL2 and TP73—the secondary line of RB1/TP53 family-mediated tumor suppression
The presence of functional homologs may underlie the long tumor latency following loss of RB and P53. RBL1 and RBL2 (also known as p107 and p130, respectively) are structurally and functionally related to RB, and although they have distinct functions in some contexts, they also regulate the cell cycle (25-30). Mutations in RBL1 and RBL2 have been identified in 13% of human SCLC (4,31). Impaired expression of RBL2 detected in patient-derived SCLC cell lines resulted from a single point mutation in the splice acceptor sequence of the first intron (31). Aberrant methylation at the promoter/regulatory region of RBL2 resulted in the gene silencing in SCLC tumors (32). These observations led to the hypothesis that RBL2 serves as a secondary tumor suppressor during SCLC development. This was validated in a recent study in which Rb/p53-mutant GEMM with homozygous or heterozygous deletion of Rbl2 showed significantly higher tumor incidence and shorter tumor latency than those with wild type (19). Notably, the dose-dependent phenotype of Rbl2-mutant mice is supported by the reduced Rbl2 transcript level in Rb/p53 tumors (19).
Similar to how RBL2 provides redundancy in RB-deficient cells, the P53 homologs TP73 and TP63 cause cell cycle arrest and apoptosis through their ability to activate expression of P53-target genes (33-37). These members of the P53 family frequently exist in multiple isoforms, including those with truncated N-terminal domains. These N-terminal truncated forms, namely ∆NP73 and ∆NP63, are products of transcripts expressed from alternative promoters (38-41). Lacking the transactivation domain but retaining the domains for DNA binding and oligomerization, these truncated forms have dominant-negative activity that inhibits wild-type members of the P53 family (41-43). While ∆NP63 has not been detected in SCLC, a recent study uncovered genomic breakpoints or mutations in the P73 locus in 13% of patient samples (n=110). Some genomic rearrangements are expected to result in the N-terminal truncated variants of P73 (designated as P73∆ex2, P73∆ex2/3) (4). These variants may have dominant negative activity on wild-type P73 and P53 as previously shown (41,44). Their roles in SCLC progression remain unknown and are being determined using the preSC-based tumor model in which similar variants are generated using CRISPR-mediated deletion of target exons. This model, once developed, will also provide a robust system to test therapeutics targeting P73-dependent tumor growth (44).
MYC family—driving early transformation and defining molecular subsets of SCLC
The MYC family of transcription factors, including MYC (c-MYC), MYCL (L-MYC), and MYCN (N-MYC), are frequently amplified and overexpressed in both SCLC cell lines and patient tumors (4,45-49). While genomic amplification of the MYC genes is detected in a significant portion (6–24%) of the patient tumors (4,46,50), it is more prevalent (32–44%) in SCLC cell lines (46,49,51). The amplification of each MYC family member occurs in a mutually exclusive manner, indicating functional redundancy among the family members in their contribution to SCLC tumorigenesis (48,49). A large body of evidence demonstrates that the MYC proteins promote tumor progression by deregulating oncogenic transcription, cell cycle control, and metabolism (21,52). However, exact roles for MYC proteins and their temporal specificity remain far from being well understood (21,53-55). For instance, almost 30 years after its discovery in SCLC, the role of L-MYC was formally tested in recent studies in which retroviral L-MYC amplification in the Rb/p53-mutant mice and Rb/p53-mutant preSC enhanced tumor development and caused transformation, respectively (21,53,56). Conversely, deletion of L-Myc in two variants of the SCLC GEMM (p53lox/lox; Rblox/lox; Rbl2lox/lox and p53lox/lox; Rblox/lox; Ptenlox/lox) suppressed tumor incidence and burden in the affected lungs (21). Furthermore, enhanced ribosome biogenesis and protein synthesis were the most significant molecular alterations during L-Myc-driven transformation of the preSCs. Inhibition of the ribosome biogenesis using a specific inhibitor, CX-5461, was shown to suppress tumor growth in the Rb/p53-mutant GEMM (21).
Similar to the L-MYC studies above, an in vivo model of c-MYC-driven SCLC has also been developed using another variant of SCLC GEMM (Rblox/lox; p53lox/lox; MycLSL/+) in which conditional expression of c-Myc drives development of tumors with a higher capacity for proliferation and metastasis than any other GEMMs, including the Rb/p53-mutant GEMM (Rblox/lox; p53lox/lox) (54). Notably, these MYC-driven tumors showed increased expression of a neurogenic transcription factor, NeuroD1, but diminished expression of another neurogenic transcription factor, Ascl1. This expression pattern coincided with reduced expression of other NE markers including SYP and CGRP; this genetic association with MYC status is also observed in both SCLC tumors and cell lines (54). Furthermore, the MYC-driven tumors in the GEMM and human SCLC with high MYC expression both displayed specific (selective) sensitivity to Aurora kinase inhibition; treatment with alisertib (a small molecule inhibitor of Aurora A kinase) drastically suppressed the growth of mouse and human MYC-driven SCLC that otherwise rapidly relapsed following standard chemotherapy, e.g., single dose of cisplatin or etoposide, or combination of both treatment (54). Taken together, these findings from the study of the GEMMs and the novel preSC-based model have helped define molecular subtypes of MYC family-driven SCLC, uncovering the oncogenic mechanisms and vulnerabilities unique to each member of the family.
Transcription factors for neural differentiation and tumor heterogeneity
The NE features of SCLC formed the basis of the idea that pulmonary neuroendocrine cells (PNECs) are the cell-of-origin, which was proven in the studies of the Rb/p53-mutant GEMM (57-59). The NE characteristics of SCLC also led numerous efforts to determine roles for NE-lineage transcription regulators in the pathogenesis of SCLC. These lineage regulators include ASCL1, NEUROD1, SOX2, TTF-1, BRN2, INSM1, and GFI1/1B to name a few. Recent studies suggest that, in addition to functioning as lineage-survival oncogenes, these factors define molecular subsets of SCLC cells and contribute to intra-tumoral heterogeneity.
ASCL1 and NEUROD1—regulators of NE differentiation and subtypes of SCLC
ASCL1 (achaete-scute-like 1; also known as HASH1) is a member of the basic helix-loop-helix (bHLH) transcription factor that has roles in neuronal commitment and differentiation of cells including PNECs during development. It also regulates stemness, cell cycle progression, and mitosis (60-62). Unlike its widespread expression among PNECs in the fetal lung, ASCL1 expression is limited to a subpopulation of PNECs—potentially dormant progenitor cells—in mature lung and is increased in subsets of high-grade NE tumors, including SCLC (63,64). ASCL1 was shown to be essential for maintaining the survival of SCLC cell lines and also required for the tumor development in GEMMs (65-67). Likewise, NEUROD1, another bHLH transcription factor, has critical roles in promoting neurogenic differentiation of cells during development and malignant behavior in SCLC cell lines (68). NEUROD1 expression is limited to a subset of SCLC cell lines distinct from those expressing ASCL1 (67,69). Intriguingly, however, NEUROD1 is not essential for the development of mouse SCLC that absolutely depends on ASCL1 expression (67). Stratification of SCLC based on differential expression patterns of ASCL1 and NEUROD1 reveals subtypes of ASCL1High (70%), NEUROD1High (15%), and ASCL1Low/NEUROD1Low (15%) and indicates that the SCLC developed in Rb/p53-mutant GEMM may resemble more the ASCL1High subtype (67). These subtypes also express distinct sets of SCLC-related oncogenes, among which L-MYC, RET, SOX2, and NFIB are targets of ASCL1 while c-MYC is a target of NEUROD1. Notably, the ASCL1High subtype tends to express L-MYC and NEUROD1High subtype express c-MYC, indicating genetic relationship between ASCL1/NEUROD1 and L-MYC/MYC (21,54). Therefore, it will be interesting to test whether these ASCL1/NEUROD1 subtypes display similar vulnerabilities to ribosome biogenesis inhibition/Aurora kinase inhibition as seen in L-MYC/c-MYC subtypes do (21,49,54).
SOX2, TTF-1, BRN2, INSM1, GFI1—potential lineage factors for cellular homeostasis
SOX family of transcription factors are involved in various aspects of morphological determination during embryonic development and also in the specification and self-renewal of tissue stem/progenitor cells (70). Several members of the B and C groups of the SOX family, including SOX2 and SOX4/11, have been detected in SCLC. SOX2 is frequently amplified and over-expressed in SCLC tumors (~27%) and cell lines (71). A high titer of SOX2 is also detected in circulation, indicating that antibodies against these factors could serve as specific serological markers for detection of SCLC (72-74). The role of SOX2 is not known, although it has been proposed as a lineage-survival oncogene (75). Expression of SOX4 target genes is increased in lung tumors with SOX4 gene overexpression (76). Therefore, SOX family transcription factors may have important roles in the expression of various genes through communication with cells present in the surrounding microenvironment.
TTF-1 (thyroid transcription factor 1; also known as NKX2-1) is one of the master regulators of epithelial differentiation and branching morphogenesis during lung development (77,78). The observation of widespread TTF-1 expression in SCLC (almost 90% of patient tumors) led to the hypothesis that it functions as a lineage survival oncogene in SCLC as it does for lung adenocarcinoma (79). Additionally, TTF-1 expression was associated with improved response to chemotherapy treatment (80). The majority of TTF-1-positive SCLCs were found at the lung periphery, and this peripheral-type SCLC had a worse prognosis than centrally located tumors (80). Our current understanding of the role of TTF-1 in SCLC is not sufficient to determine whether its expression and activity are reliable biomarkers for prognosis and therapy, necessitating more functional and mechanistic studies.
BRN2 (brain-2; also known as POU3F2) is a neural cell-specific POU domain transcription factor for neural lineage determination (81). An ectopic expression and knockdown experiment suggested that BRN2 functions upstream of ASCL1 and NEUROD1 to promote expression of NE genes and promote cell proliferation (82). RB may be an upstream regulator of BRN2 as RB-mutant retinoblastoma expresses high levels of BRN2 which are reduced with restoration of RB expression (83). Additionally, the BRAF-mediated activation of BRN2 in melanoma could provide insight into this interaction in SCLC because BRAF mutation, although rare, has been discovered in SCLC patient tumor (4,84).
INSM1 (insulinoma-associated 1) is a transcription factor with zinc-finger DNA binding domain and a SNAG (SNAIL/GFI1)-domain that was originally isolated from a human insulinoma. It plays an important role in the development of NE cells in the pancreas and intestines, and adrenal medulla and basal neuronal progenitor cells in the neocortex (85-88). Insm1 binds directly to regulatory sequences in the Hes1 gene to repress its expression, Mutation of Insm1 leads to upregulated Hes1 expression and interferes with maintenance of Ascl1 expression (89). Conversely, NOTCH/HES1 signaling suppresses INSM1 (90). The observation of its expression in nearly all SCLC cells prompted the hypothesis that it is a crucial regulator of NE differentiation in this cancer (91-93). Knockdown of INSM1 reduced proliferation rates of SCLC cell lines and expression of neuroendocrine-specific genes, including ASCL1, BRN2, CHGA, SYP, and NCAM (90).
GFI1 (growth factor independent-1) and its homolog GFI1B, proteins with zinc-finger DNA binding domains and SNAG domains, are transcriptional repressors critical for development of the hematopoietic system (94,95). GFI1/1B, acting downstream of proneural bHLH factors such as ASCL1 and MATH1, also plays a role in NE differentiation and SCLC development (96-98). GFI1 knockout drastically reduces NE differentiation and impairs PNEC proliferation (97,99). Mechanistically, GFI1 binds to regulatory regions of target genes and recruit’s chromatin modifiers, including LSD1, that in turn demethylase H3K4 to repress gene expression (100). This interaction likely explains the effect of LSD1 inhibitors on SCLC growth, together with the finding that one of the inhibitors interfered with the interaction of LSD1 with GFI1B and INSM1, another SNAG-containing protein (101,102).
Signaling pathways to transcription effectors for intratumoral heterogeneity
Recently, studies have determined the cellular and molecular origin of the intratumoral heterogeneity seen in SCLC. NOTCH singling plays an important role in cell-fate decisions in a variety of tissues (103). During lung development, a subset of epithelial progenitor cells expressing Delta-like ligands (DLLs) inhibits NE differentiation of adjacent progenitor cells expressing NOTCH via DLL-NOTCH interaction-mediated signaling that suppresses ASCL1 expression in the NOTCH-expressing cells (104). However, much less is known about the role of the NOTCH signaling in the differentiation of SCLC that consists mainly of DLLs-expressing NE cells but only few other type of epithelial cells expressing NOTCH receptors. Somatic mutations affecting genes encoding NOTCH 1, 2, 3 and 4 are detected in 25% of patient tumors, and the majority of cases also had high levels of DLK1 (delta-like 1 homolog), an inhibitor of Notch signaling (4,105). Activation of NOTCH signaling through expression of N1ICD/N2ICD, the transcriptional effectors, inhibits cell cycle progression in SCLC cells and reduces tumor development in the lungs of Rb/p53-mutant GEMM (4,106). These data, while suggesting a tumor suppressive role for the NOTCH pathway, do not readily support activation of the pathway as a viable therapeutic strategy, in part because this pathway acts tumor suppressive and oncogenic in different populations of SCLC (107).
Using NE cell-specific adeno-CGRP-Cre and a Hes1GFP/+ allele (a knock-in reporter of NOTCH activity) with SCLC GEMM, a study found that NOTCH/HES1-active GFPpositive cells coexisted with NOTCH/HES1-inactive GFPnegative cells originating from the NE cells (107). The HES1-active cells express known target genes of the pathway, including Notch genes themselves and Nrarp, while suppressing NE genes. This NOTCH/HES1-driven switch to non-NE phenotype occurred in 10–50% of NE cells and coincided with induction of REST, a transcriptional repressor and a direct target of NOTCH pathway that represses expression of NE genes. On the other hand, the HES1-negative NE cells expressed ligands, DLL1, 3, and 4 and NE genes typical of SCLC. Additionally, the HES1-active non-NE cells were relatively resistant to carboplatin and irinotecan at doses that effectively killed the HES1-inactive NE cells. These findings led to a model in which the non-NE SCLC cells promote malignant progression and facilitate regeneration of NE cells following chemotherapies, suggesting that Notch inhibition in combination with chemotherapy may be more efficacious in preventing early-stage SCLC progression or relapse following existing chemotherapies. Furthermore, given the NOTCH/HES1-driven regulation of ASCL1 and L-MYC/c-MYC, it is possible that the pathway may play a role in defining ASCL1High/NEUROD1High/double-negative subtypes and/or L-MYC/c-MYC subtypes of SCLC cells described above (66,106,108) and that these molecular subtypes would have differential sensitivities to a combination of NOTCH inhibition and conventional chemotherapy (54). These findings have advanced our understanding of intratumoral heterogeneity in SCLC since the initial model that proposed a role of non-NE cells in metastasis of NE cells (109).
Signaling pathways that contribute to SCLC heterogeneity via transcriptional change are likely complex, including more pathways implicated in normal lung development and homeostasis and cross talks among them. For instance, PEA3, a member of the ETS transcription factor family, has been implicated in maintaining the intratumoral heterogeneity (110). High expression of PEA3 was detected in NE cells in tumor that were treated with conditioned medium from non-NE cells. The increased PEA3 was sufficient to induce the invasive migration property in NE cells. Fibroblast growth factor (FGF)/RAS/MAPK pathway regulated the PEA3 expression and invasiveness in NE cells, suggesting that the FGF pathway is responsible for paracrine signaling between NE and non-NE cells in SCLC. These findings are in line with previous studies that showed that PEA3 was expressed in metastatic tumors, and that its expression correlated with metastasis of various human cancers, including breast cancer and NSCLC (111-113), implying a role of PEA in paracrine signaling in the lung tumor microenvironment. In light of these roles of signaling pathways initiated by membrane-bound receptors, it is also worth revisiting the Hedgehog (HH) signaling in the context of SCLC intratumoral heterogeneity. Similar to NOTCH and FGFR pathways, HH signaling is transmitted to activate the GLI family of zinc-finger transcription factors that induce numerous oncogenes including c-MYC, CCND1, and the GLIs themselves (114). Preclinical studies have shown that an autocrine, ligand-dependent signaling promotes SCLC development, while other studies showed a significance of a paracrine signaling to tumor-associated stroma cells (115-118). While it remains debatable, to what extent these modes of signaling contribute to the pathogenesis of SCLC, the activated GLI promotes proliferation and cell-cell interactions. It is tempting to speculate whether non-NE cells described above are HH-responsive stroma cells and the transcriptional activity of GLI contributes the development and maintenance of tumor heterogeneity. Better understanding of HH signaling in this emerging context of SCLC would provide novel insight into targeted therapy against the signaling (119).
Epigenetic alterations driving malignant progression
Discovery of aberrant epigenetic drivers of SCLC progression has been an important step toward understanding the SCLC biology. Recent studies have uncovered a novel function of NFIB in regulating chromatin states for metastasis and have begun to unravel the complex network of chromatin modifiers, including histone methyltransferases, demethylases, and acetyltransferases, and other components of multi-protein histone modifying complexes.
NFIB altering chromatin states for SCLC metastasis
NFIB, a member of the nuclear factor I (NFI) family of transcription factors, binds to promoter, enhancer, and silencer regions in the genome and regulates a plethora of genes in almost all tissues during development (120,121). A study of Nfib knockout mice showing developmental defects in lung and brain supports a fundamental role for the gene in a wide range of biological processes (122,123). NFIB alterations have been implicated in malignancies; particularly, it is amplified in human SCLC tumors (15%), cell lines (34%) and mouse SCLC developed in the Rb/p53-mutant GEMM (124,125). Nfib knockdown reduced cell proliferation while increasing cell death, suggesting a role for the gene in maintaining tumor cell homeostasis (124). Recently, a novel role of NFIB and its in vivo relevance have been determined in three independent studies using variants of the SCLC GEMM (125-127). These studies found that (I) Nfib amplification and overexpression are more prevalent in metastases than primary tumors; (II) ectopic Nfib expression accelerated mouse SCLC progression with increased tumor burden and metastasis; and (III) increased Nfib was both sufficient and required for multiple steps of metastasis to lymph nodes and liver from primary tumors. Furthermore, ATAC-seq (assay of transposase-accessible chromatin with sequencing) and lung tumors and liver metastasis from the same GEMM showed that Nfib, once bound to target DNA elements, initiates and stabilizes accessible chromatin configuration that promotes expression of genes required for metastatic progression of mouse SCLC cells (124,126). These Nfib-induced changes in chromatin accessibilities genome-wide coincide with altered expression of a large number of genes functionally related to neural development/differentiation, cell adhesion, and motility (124,126). Transcriptional analyses in the other studies revealed that Nfib regulates the expression of genes related to axon guidance, focal adhesion and extracellular matrix-receptor interactions, and cellular movement (125,127). High levels of Nfib are associated with expansive growth of a poorly differentiated and E-cadherin (CDH1)-negative invasive population of tumor cells, which corresponds to features of stage III/IV high-grade NE carcinomas in patients (125). These findings strongly suggest that concerted actions of Nifb target genes drive metastasis of SCLC mainly by altering cell-adhesion and movement. NFIB and its mechanism of action in tumor development and metastasis may present viable targets for intervening in tumor development and metastasis.
EZH2 (Enhancer of zeste homolog 2)—epigenetic regulator for malignant progression
EZH2 is a histone methyltransferase that, together with EED and SUZ12, forms the polycomb repressive complex 2 (PRC2) (128). It mediates tri-methylation of histone H3 at lysine 27 (H3K27me3) in discrete promoter CpG islands, leading to transcriptional repression (128,129). In addition to its role in promoting heterochromatin formation and gene silencing during development and differentiation, increased EZH2 expression has been linked to numerous cancer types including lung cancers (130-132). In cancer cells, high EZH2 activity results in the long-term repression of tumor suppressor genes (133). Remarkably, ectopic expression of EZH2 alone was sufficient to cause NSCLC in mice (134). EZH2 overexpression also promotes development of K-Ras-driven NSCLC (135). In SCLC cell lines, EZH2 levels are expressed three times greater than those in NSCLC lines and twelve times greater than in normal lungs (132). Since EZH2 is a known target of E2F transcription factor, complete loss of RB in SCLC likely results in deregulated EZH2 expression (129,132). Overexpression of EED and SUZ12 coincided with high EZH2 overexpressed in SCLC tumor samples, further indicating the presence of the PRC2 complex (136). Functionally, EZH2 plays a role in homeostasis of SCLC cells as its knockdown increased apoptotic activity by up-regulating the pro-apoptotic factors such as PUMA and BAD and by elevating P21 protein levels (137). ChIP-seq analysis indicated that JUB (AJUBA) is the most repressed gene of hyper-methylated H3K27me3 in SCLC cell lines (136). Notably, the extents of JUB gene repression correlated with reduced patient survival, suggesting role of the PRC2 activity is linked with poor SCLC prognosis (136,138).
Given its widespread involvement in malignancy, there has been considerable interest in reversing EZH2-mediated repression of tumor suppressor genes. Recent efforts to test small molecule inhibitors of EZH2 in lung cancer have met with some success. EZH2 inhibition using GSK126 and DNZep sensitized BRG1 and EGFR-mutant lung adenocarcinoma to existing chemotherapy (139). Another inhibitor, JQEZ5, also suppressed growth of EZH2-driven/addicted lung tumors in GEMMs and xenografts of human NSCLC cell lines (134). Likewise, inhibition of EZH2 using EPZ-6438, suppressed growth of patient-derived xenograft as well as SCLC cells in culture (140). These results strongly support EZH2 inhibition as strategy for intervening in SCLC progression. However, it may be challenging to stratify SCLC tumors for this EZH2-targted therapy, as EZH2 expression alone may not be sufficient for SCLC development. Identification of EZH2-targets would help identify reliable biomarker to predict the efficacy of EZH2 inhibitors. One potential biomarker for EZH2 activity is SLFN11 (Schlafen family member 11), a putative DNA/RNA helicase whose expression could sensitize cells to DNA-damaging agents (141-144). SLFN11 expression was diminished by EZH2-driven methylation in SCLC cells treated with talazoparib, a PARP inhibitor, and inactivation of SLFN11 using shRNA or CRISPR/Cas9 conferred resistance to the drug (145). These findings indicated SLFN11 as a predictive biomarker of sensitivity to PARP-targeted therapy in SCLC and led to a preclinical study in which EZH2 inhibition in combination with PARP inhibition restored SLFN11, thereby restoring chemo-sensitivity.
MLL family of histone methyltransferases
The MLL (mixed lineage leukemia; also known as KMT2) proteins as part of multi-protein complexes, regulate methylation of lysine 4 and 27 residues on histone H3 tails (H3K4 and H3K27) in regulatory elements of genes (146). MLL1/4 (KMT2A/B) complex methylates H3K4 and MLL2/3 (KMT2D/C)-UTX (KDM6A) complex demethylases H3K27 (146,147). Initially discovered in hematopoietic malignancies with SET domain deletion leading to hypomethylation of H3K4 and transcriptional inactivation, MLL family mutations are among the most frequent alterations in cancer (148-150). In SCLC, multiple types of mutations, including missense mutations and truncations, were discovered in the genes encoding the MLLs; a majority of the mutations were truncating mutations, and MLL1 (KMT2A) and MLL2 (KMT2D) were the most frequently mutated in both SCLC cell lines and the patient tumors (48,71,151-153). These mutations were associated with low protein levels and global reductions in mono-methylation of histone H3 lysine 4 (H3K4me1), a chromatin marker of transcriptional enhancers (152). A few SCLC cells with normal MLL2 mutation instead had truncating mutations in UTX with similar defects in H3K4 methylation (152). This reduced H3K4me1, given its antagonistic relationship with H3K27 methylation, may have increased H3K27 methylation driven by PRC2/EZH2 that results in gene silencing. It remains unknown how these epigenetic alterations caused by loss-of-function mutations influence tumorigenesis.
LSD1—H3K4 demethylase
LSD1 (lysine-specific demethylase 1; also known as KDM1A) demethylases mono- and di-methylated lysine 4 of histone H3 (H3K4me1/2), thereby epigenetically regulating the activation or repression of gene expression in different contexts (154-156). LSD1 is overexpressed in hematologic malignancies and solid tumors including SCLC (157-161). Two independent drug screening studies recently found that two small molecule inhibitors of LSD1, designated GSK2879552 and T-3775440, had antitumor effects on SCLC cells in vitro and in vivo (101,102). Mechanistically, GSK2879552 caused widespread hypomethylation that altered expression of a number of genes including ZEB1 and IGFBP2 (101) and T-3775440 disrupted the interaction between LSD1 and the SNAG (SNAIL/GFI1) domain transcription factors INSM1 and GFI1B, thereby inhibiting expression of NE-associated genes, such as ASCL1 (102,162). The mechanism of LSD1 action in SCLC remains to be determined; demethylation of H3K4 on enhancer regions of tumor suppressor genes may cause transcription inhibition and modify the histones near MYC binding sites to promote its transcription activity (163). Elucidation of LSD1-mediated transcriptional regulation will provide important insight into a novel therapeutic strategy involving inhibition of LSD1, which is being tested in a clinical trial.
CREBBP/EP300 family of acetyltransferases
CREBBP [cAMP response element-binding (CREB)-binding protein] and EP300 (E1A associated p300) have intrinsic histone acetyl transferase (HAT) activity and play critical roles in embryonic development, growth control, and homeostasis by coupling chromatin remodeling to transcription factor recognition (164-166). Somatic mutations in these homologous factors were found in multiple cancer types, including lung cancers (48,167,168). In SCLC, a significant fraction of patient tumors carries mutations in the genes encoding these factors (4,48,71,152). The clustering pattern of missense mutations in the exons encoding the HAT domain indicates significance of the catalytic function for tumor suppression and, together with mutual exclusiveness between CREBBP and EP300 mutations, also suggests that those affecting the HAT domain may have dominant-negative functions on wild-type proteins and functional paralog (4). The functional significance of these mutant forms of CREBBP/EP300 in SCLC is currently being determined using the Rb/p53-mutant GEMMs as well as the preSCs. It is expected that removing Crebbp or Ep300 from mouse lung epithelium already lacking Rb and p53 will accelerate SCLC development. CRISPR-mediated mutation of the HAT domain, resulting in its truncation, is expected to cause transformation of the preSCs. These studies will support a tumor-suppressive role for these HAT-containing transcription co-factors.
Mechanistically, CREBBP/EP300 acetylates H3K27 in the enhancer regions of target genes throughout the genome to promote transcription which, in concert with the MLL3/4-UTX demethylase complex, opposes the PRC2-mediated methylation of the histones that usually represses gene expression (169-171). In the context of tumor cells with altered CREBBP/EP300 activities, modification of H3K27 is perturbed and the affected genes, likely tumor suppressors, are highly methylated and suppressed. Identification of these suppressed genes will enhance our understanding of the tumorigenesis driven by these alterations. However, a different mechanism of CREBBP/EP300 actions explains their regulation of oncogenes including MYC. It has been reported that CREBBP-deficient cancer cell lines and CREBBP-knockout cells are uniquely susceptible to EP300 depletion. Inactivation of both CREBBP and EP300 enhances the H3K27 methylation but also cause decreased expression of MYC; the latter turns out to be detrimental to the NSCLC cells (172). Therefore, the mutually exclusive pattern of CREBBP and EP300 mutation may suggest not only functional redundancy of these paralogs but also a synthetic lethal relationship. Given the mutually exclusive pattern of these alterations also present in SCLC, it is critical to test the CREBBP-EP300 synthetic lethality relationship.
Other proteins in chromatin modifying complexes
Components of the BAF-SWI/SNF and PBAF-SWI/SNF complexes are implicated in SCLC (152). Mutations have been found in ARID1A/B (AT-rich interaction domain 1 A and B), PBRM1 (polybromo 1; also known as BAF180), and BRG1 (SMARCA4). CHD7 (chromodomain helicase DNA binding protein 7), known to interact with the PBRM1-containing PBAF complex, is also mutated in SCLC. Collectively, these mutations are found in significant portions of SCLC, suggesting a role for SWI/SNF complexes-mediated epigenetic regulation in SCLC development. While the functional significance of these mutations remains to be determined, their known function and relationships with other oncogenic factors have been exploited to devise therapeutic strategies. For example, in line with the opposing functions of BAF complexes and the PRC2 complex, cancers with ARID1A loss are highly sensitive to inhibition of EZH2 in the PRC2 (173). Also, the additional function of ARID1A in response to DNA double-strand DNA breaks (DSBs) supports the concept that loss of ARID1A sensitizes tumor cells to DSB-inducing radiation or PARP inhibitors. Thus, ARID1A mutation may a reliable biomarker for treatment with a PARP inhibitor shown to reduce tumor growth in preclinical models (174). BRG1 regulates expression of MAX, the dimerization partner of MYC, and cooperates with MYC/MAX in expression of a common set of genes in SCLC. But depletion of BRG1 causes lethality specifically in MAX-deficient cell by significantly affecting MYC in gene expression (175). Additionally, BRD4 (Bromodomain-containing protein 4), a protein that binds to acetylated histones and recruit’s chromatin modifiers and transcription factors, has been a molecular target of interest since a bromodomain inhibitor JQ1 inhibited the growth of cancer cells with a significantly higher efficacy in MYC-amplified SCLC lines (176,177). Although not altered in SCLC, BRD4 may be a rational target, given its role in expression of MYC and other oncogenes (178). Other reports, however, show that BRD4 occupies the enhancer region of ASCL1 and JQ1 inhibits expression of ASCL1, not MYC, in SCLC cells (179) and that the sensitivity to JQ1 does not correlate with the levels of L-MYC, N-MYC, and ASCL1 but rather with CDK6 (180). These different results reflect cell context-specific BRD4 occupation in the regulatory regions of target genes.
Conclusions
Propelled by the discovery of genomic alterations and advances in models and analytical methods, the field of SCLC research has seen remarkable progress in understanding tumor development and malignant progression and developing new strategies for treatment. However, only a few of the altered transcription regulators have been evaluated for their roles in the tumor development and in vivo relevance. In addition to determining the roles of individual factors altered in the cancer genome, understanding functional relationships among them will lead to a model of transcriptional pathways that converge on regulatory regions of crucial genes during tumor development. Upregulated transcription factors such as MYCs and NFIB occupy the regulatory regions of oncogenes and recruit histone modifiers such as LSD1 to enhance transcription. Repressor complexes such as EZH2-containing PRC2 methylate H3K27 at the regulatory regions of tumor suppressor genes. Resulting gene expression changes favor oncogenic progression that is further amplified by loss-of-function alterations in MLL1/2, MLL3/4-UTX complex, and CREBBP/EP300, which normally antagonizes the PRC2/EZH2-mediated silencing of target genes (Figure 1C). Validation of this model and characterization of target genes will move the field forward to define oncogenic mechanisms and vulnerabilities common in molecular subtypes and provide critical insights into novel strategies for tumor intervention.
Acknowledgements
We would like to thank Colin Dunn for helpful comments on the manuscript.
Funding: This work is funded in part by NIH/NCI (R01CA194461 and R03CA215777), American Cancer Society (RSG-15-066-01-TBG), DOD/CDMRP (Concept Award, LC140542), and the David R. Jones Fund (UVA Dept. Surgery) to KS Park and National Research Foundation of Korea (2016R1D1A3B02006754) and Basic Research Program of Kangwon National University to KC Kim.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Takahashi T, Nau MM, Chiba I, et al. p53: a frequent target for genetic abnormalities in lung cancer. Science 1989;246:491-4. [Crossref] [PubMed]
- Hensel CH, Hsieh CL, Gazdar AF, et al. Altered structure and expression of the human retinoblastoma susceptibility gene in small cell lung cancer. Cancer Res 1990;50:3067-72. [PubMed]
- Mori N, Yokota J, Akiyama T, et al. Variable mutations of the RB gene in small-cell lung carcinoma. Oncogene 1990;5:1713-7. [PubMed]
- George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47-53. [Crossref] [PubMed]
- Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell 2002;2:103-12. [Crossref] [PubMed]
- Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer 2008;8:671-82. [Crossref] [PubMed]
- Zilfou JT, Lowe SW. Tumor suppressive functions of p53. Cold Spring Harb Perspect Biol 2009;1:a001883. [Crossref] [PubMed]
- Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol 2013;15:2-8. [Crossref] [PubMed]
- Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 2014;25:304-17. [Crossref] [PubMed]
- Vélez-Cruz R, Johnson DG. The retinoblastoma (RB) tumor suppressor: pushing back against genome instability on multiple fronts. Int J Mol Sci 2017;18:E1776. [Crossref] [PubMed]
- Vogel F. Genetics of retinoblastoma. Hum Genet 1979;52:1-54. [Crossref] [PubMed]
- Friend SH, Dryja TP, Weinberg RA. Oncogenes and tumor-suppressing genes. N Engl J Med 1988;318:618-22. [Crossref] [PubMed]
- Gouyer V, Gazzeri S, Bolon I, et al. Mechanism of retinoblastoma gene inactivation in the spectrum of neuroendocrine lung tumors. Am J Respir Cell Mol Biol 1998;18:188-96. [Crossref] [PubMed]
- Sekido Y, Fong KM, Minna JD. Molecular genetics of lung cancer. Annu Rev Med 2003;54:73-87. [Crossref] [PubMed]
- Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer 2006;6:909-23. [Crossref] [PubMed]
- Carvajal LA, Manfredi JJ. Another fork in the road--life or death decisions by the tumour suppressor p53. EMBO Rep 2013;14:414-21. [Crossref] [PubMed]
- Harms K, Nozell S, Chen X. The common and distinct target genes of the p53 family transcription factors. Cell Mol Life Sci 2004;61:822-42. [Crossref] [PubMed]
- Meuwissen R, Linn SC, Linnoila RI, et al. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell 2003;4:181-9. [Crossref] [PubMed]
- Schaffer BE, Park KS, Yiu G, et al. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer Res 2010;70:3877-83. [Crossref] [PubMed]
- Wikenheiser-Brokamp KA. Rb family proteins differentially regulate distinct cell lineages during epithelial development. Development 2004;131:4299-310. [Crossref] [PubMed]
- Kim DW, Wu N, Kim YC, et al. Genetic requirement for Mycl and efficacy of RNA Pol I inhibition in mouse models of small cell lung cancer. Genes Dev 2016;30:1289-99. [Crossref] [PubMed]
- Roth JA, Nguyen D, Lawrence DD, et al. Retrovirus-mediated wild-type p53 gene transfer to tumors of patients with lung cancer. Nat Med 1996;2:985-91. [Crossref] [PubMed]
- Feldser DM, Kostova KK, Winslow MM, et al. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature 2010;468:572-5. [Crossref] [PubMed]
- Junttila MR, Karnezis AN, Garcia D, et al. Selective activation of p53-mediated tumour suppression in high-grade tumours. Nature 2010;468:567-71. [Crossref] [PubMed]
- Zhu L, van den Heuvel S, Helin K, et al. Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein. Genes Dev 1993;7:1111-25. [Crossref] [PubMed]
- Claudio PP, Howard CM, Baldi A, et al. p130/pRb2 has growth suppressive properties similar to yet distinctive from those of retinoblastoma family members pRb and p107. Cancer Res 1994;54:5556-60. [PubMed]
- Baldi A, De Luca A, Claudio PP, et al. The RB2/p130 gene product is a nuclear protein whose phosphorylation is cell cycle regulated. J Cell Biochem 1995;59:402-8. [Crossref] [PubMed]
- Zhu L, Enders G, Lees JA, et al. The pRB-related protein p107 contains two growth suppression domains: independent interactions with E2F and cyclin/cdk complexes. EMBO J 1995;14:1904-13. [PubMed]
- Claudio PP, De Luca A, Howard CM, et al. Functional analysis of pRb2/p130 interaction with cyclins. Cancer Res 1996;56:2003-8. [PubMed]
- Caputi M, Groeger AM, Esposito V, et al. Loss of pRb2/p130 expression is associated with unfavorable clinical outcome in lung cancer. Clin Cancer Res 2002;8:3850-6. [PubMed]
- Helin K, Holm K, Niebuhr A, et al. Loss of the retinoblastoma protein-related p130 protein in small cell lung carcinoma. Proc Natl Acad Sci U S A 1997;94:6933-8. [Crossref] [PubMed]
- Cinti C, Macaluso M, Giordano A. Tumor-specific exon 1 mutations could be the 'hit event' predisposing Rb2/p130 gene to epigenetic silencing in lung cancer. Oncogene 2005;24:5821-6. [Crossref] [PubMed]
- Kaghad M, Bonnet H, Yang A, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 1997;90:809-19. [Crossref] [PubMed]
- De Laurenzi V, Costanzo A, Barcaroli D, et al. Two new p73 splice variants, gamma and delta, with different transcriptional activity. J Exp Med 1998;188:1763-8. [Crossref] [PubMed]
- Ueda Y, Hijikata M, Takagi S, et al. Transcriptional activities of p73 splicing variants are regulated by inter-variant association. Biochem J 2001;356:859-66. [Crossref] [PubMed]
- Costanzo A, Merlo P, Pediconi N, et al. DNA damage-dependent acetylation of p73 dictates the selective activation of apoptotic target genes. Mol Cell 2002;9:175-86. [Crossref] [PubMed]
- Flores ER, Tsai KY, Crowley D, et al. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 2002;416:560-4. [Crossref] [PubMed]
- Yang A, McKeon F. P63 and P73: P53 mimics, menaces and more. Nat Rev Mol Cell Biol 2000;1:199-207. [Crossref] [PubMed]
- Pozniak CD, Radinovic S, Yang A, et al. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science 2000;289:304-6. [Crossref] [PubMed]
- Sayan AE, Sayan BS, Findikli N, et al. Acquired expression of transcriptionally active p73 in hepatocellular carcinoma cells. Oncogene 2001;20:5111-7. [Crossref] [PubMed]
- Tannapfel A, John K, Mise N, et al. Autonomous growth and hepatocarcinogenesis in transgenic mice expressing the p53 family inhibitor DNp73. Carcinogenesis 2008;29:211-8. [Crossref] [PubMed]
- Stiewe T, Putzer BM. Role of p73 in malignancy: tumor suppressor or oncogene? Cell Death Differ 2002;9:237-45. [Crossref] [PubMed]
- Jancalek R. The role of the TP73 gene and its transcripts in neuro-oncology. Br J Neurosurg 2014;28:598-605. [Crossref] [PubMed]
- Venkatanarayan A, Raulji P, Norton W, et al. IAPP-driven metabolic reprogramming induces regression of p53-deficient tumours in vivo. Nature 2015;517:626-30. [Crossref] [PubMed]
- Brennan J, O'Connor T, Makuch RW, et al. myc family DNA amplification in 107 tumors and tumor cell lines from patients with small cell lung cancer treated with different combination chemotherapy regimens. Cancer Res 1991;51:1708-12. [PubMed]
- Johnson BE, Brennan JF, Ihde DC, et al. myc family DNA amplification in tumors and tumor cell lines from patients with small-cell lung cancer. J Natl Cancer Inst Monogr 1992.39-43. [PubMed]
- Johnson BE, Russell E, Simmons AM, et al. MYC family DNA amplification in 126 tumor cell lines from patients with small cell lung cancer. J Cell Biochem Suppl 1996;24:210-7. [Crossref] [PubMed]
- Peifer M, Fernandez-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104-10. [Crossref] [PubMed]
- Sos ML, Dietlein F, Peifer M, et al. A framework for identification of actionable cancer genome dependencies in small cell lung cancer. Proc Natl Acad Sci U S A 2012;109:17034-9. [Crossref] [PubMed]
- Gazzeri S, Brambilla E, Jacrot M, et al. Activation of myc gene family in human lung carcinomas and during heterotransplantation into nude mice. Cancer Res 1991;51:2566-71. [PubMed]
- Johnson BE, Ihde DC, Makuch RW, et al. myc family oncogene amplification in tumor cell lines established from small cell lung cancer patients and its relationship to clinical status and course. J Clin Invest 1987;79:1629-34. [Crossref] [PubMed]
- Dang CV. MYC on the path to cancer. Cell 2012;149:22-35. [Crossref] [PubMed]
- Huijbers IJ, Bin Ali R, Pritchard C, et al. Rapid target gene validation in complex cancer mouse models using re-derived embryonic stem cells. EMBO Mol Med 2014;6:212-25. [PubMed]
- Mollaoglu G, Guthrie MR, Bohm S, et al. MYC drives progression of small cell lung cancer to a variant neuroendocrine subtype with vulnerability to aurora kinase inhibition. Cancer Cell 2017;31:270-85. [Crossref] [PubMed]
- Semenova EA, Nagel R, Berns A. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev 2015;29:1447-62. [Crossref] [PubMed]
- Nau MM, Brooks BJ, Battey J, et al. L-myc, a new myc-related gene amplified and expressed in human small cell lung cancer. Nature 1985;318:69-73. [Crossref] [PubMed]
- Park KS, Liang MC, Raiser DM, et al. Characterization of the cell of origin for small cell lung cancer. Cell Cycle 2011;10:2806-15. [Crossref] [PubMed]
- Sutherland KD, Proost N, Brouns I, et al. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell 2011;19:754-64. [Crossref] [PubMed]
- Song H, Yao E, Lin C, et al. Functional characterization of pulmonary neuroendocrine cells in lung development, injury, and tumorigenesis. Proc Natl Acad Sci U S A 2012;109:17531-6. [Crossref] [PubMed]
- Kim HJ, McMillan E, Han F, et al. Regionally specified human neural progenitor cells derived from the mesencephalon and forebrain undergo increased neurogenesis following overexpression of ASCL1. Stem Cells 2009;27:390-8. [Crossref] [PubMed]
- Mazurier N, Parain K, Parlier D, et al. Ascl1 as a novel player in the Ptf1a transcriptional network for GABAergic cell specification in the retina. PLoS One 2014;9:e92113. [Crossref] [PubMed]
- Donakonda S, Sinha S, Dighe SN, et al. System analysis identifies distinct and common functional networks governed by transcription factor ASCL1, in glioma and small cell lung cancer. Mol Biosyst 2017;13:1481-94. [Crossref] [PubMed]
- Westerman BA, Neijenhuis S, Poutsma A, et al. Quantitative reverse transcription-polymerase chain reaction measurement of HASH1 (ASCL1), a marker for small cell lung carcinomas with neuroendocrine features. Clin Cancer Res 2002;8:1082-6. [PubMed]
- Miki M, Ball DW, Linnoila RI. Insights into the achaete-scute homolog-1 gene (hASH1) in normal and neoplastic human lung. Lung Cancer 2012;75:58-65. [Crossref] [PubMed]
- Jiang T, Collins BJ, Jin N, et al. Achaete-scute complex homologue 1 regulates tumor-initiating capacity in human small cell lung cancer. Cancer Res 2009;69:845-54. [Crossref] [PubMed]
- Augustyn A, Borromeo M, Wang T, et al. ASCL1 is a lineage oncogene providing therapeutic targets for high-grade neuroendocrine lung cancers. Proc Natl Acad Sci U S A 2014;111:14788-93. [Crossref] [PubMed]
- Borromeo MD, Savage TK, Kollipara RK, et al. ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep 2016;16:1259-72. [Crossref] [PubMed]
- Westerman BA, Breuer RH, Poutsma A, et al. Basic helix-loop-helix transcription factor profiling of lung tumors shows aberrant expression of the proneural gene atonal homolog 1 (ATOH1, HATH1, MATH1) in neuroendocrine tumors. Int J Biol Markers 2007;22:114-23. [Crossref] [PubMed]
- Osborne JK, Guerra ML, Gonzales JX, et al. NeuroD1 mediates nicotine-induced migration and invasion via regulation of the nicotinic acetylcholine receptor subunits in a subset of neural and neuroendocrine carcinomas. Mol Biol Cell 2014;25:1782-92. [Crossref] [PubMed]
- Bergsland M, Werme M, Malewicz M, et al. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev 2006;20:3475-86. [Crossref] [PubMed]
- Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 2012;44:1111-6. [Crossref] [PubMed]
- Titulaer MJ, Klooster R, Potman M, et al. SOX antibodies in small-cell lung cancer and Lambert-Eaton myasthenic syndrome: frequency and relation with survival. J Clin Oncol 2009;27:4260-7. [Crossref] [PubMed]
- Güre AO, Stockert E, Scanlan MJ, et al. Serological identification of embryonic neural proteins as highly immunogenic tumor antigens in small cell lung cancer. Proc Natl Acad Sci U S A. 2000;97:4198-203. [Crossref] [PubMed]
- Maddison P, Thorpe A, Silcocks P, et al. Autoimmunity to SOX2, clinical phenotype and survival in patients with small-cell lung cancer. Lung Cancer 2010;70:335-9. [Crossref] [PubMed]
- Bass AJ, Watanabe H, Mermel CH, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet 2009;41:1238-42. [Crossref] [PubMed]
- Castillo SD, Matheu A, Mariani N, et al. Novel transcriptional targets of the SRY-HMG box transcription factor SOX4 link its expression to the development of small cell lung cancer. Cancer Res 2012;72:176-86. [Crossref] [PubMed]
- Kimura S, Hara Y, Pineau T, et al. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev 1996;10:60-9. [Crossref] [PubMed]
- DeFelice M, Silberschmidt D, DiLauro R, et al. TTF-1 phosphorylation is required for peripheral lung morphogenesis, perinatal survival, and tissue-specific gene expression. J Biol Chem 2003;278:35574-83. [Crossref] [PubMed]
- Di Loreto C, Di Lauro V, Puglisi F, et al. Immunocytochemical expression of tissue specific transcription factor-1 in lung carcinoma. J Clin Pathol 1997;50:30-2. [Crossref] [PubMed]
- Misch D, Blum T, Boch C, et al. Value of thyroid transcription factor (TTF)-1 for diagnosis and prognosis of patients with locally advanced or metastatic small cell lung cancer. Diagn Pathol 2015;10:21. [Crossref] [PubMed]
- Nakai S, Kawano H, Yudate T, et al. The POU domain transcription factor Brn-2 is required for the determination of specific neuronal lineages in the hypothalamus of the mouse. Genes Dev 1995;9:3109-21. [Crossref] [PubMed]
- Ishii J, Sato H, Sakaeda M, et al. POU domain transcription factor BRN2 is crucial for expression of ASCL1, ND1 and neuroendocrine marker molecules and cell growth in small cell lung cancer. Pathol Int 2013;63:158-68. [Crossref] [PubMed]
- Cobrinik D, Francis RO, Abramson DH, et al. Rb induces a proliferative arrest and curtails Brn-2 expression in retinoblastoma cells. Mol Cancer 2006;5:72. [Crossref] [PubMed]
- Goodall J, Wellbrock C, Dexter TJ, et al. The Brn-2 transcription factor links activated BRAF to melanoma proliferation. Mol Cell Biol 2004;24:2923-31. [Crossref] [PubMed]
- Goto Y, De Silva MG, Toscani A, et al. A novel human insulinoma-associated cDNA, IA-1, encodes a protein with "zinc-finger" DNA-binding motifs. J Biol Chem 1992;267:15252-7. [PubMed]
- Gierl MS, Karoulias N, Wende H, et al. The zinc-finger factor Insm1 (IA-1) is essential for the development of pancreatic beta cells and intestinal endocrine cells. Genes Dev 2006;20:2465-78. [Crossref] [PubMed]
- Farkas LM, Haffner C, Giger T, et al. Insulinoma-associated 1 has a panneurogenic role and promotes the generation and expansion of basal progenitors in the developing mouse neocortex. Neuron 2008;60:40-55. [Crossref] [PubMed]
- Wildner H, Gierl MS, Strehle M, et al. Insm1 (IA-1) is a crucial component of the transcriptional network that controls differentiation of the sympatho-adrenal lineage. Development 2008;135:473-81. [Crossref] [PubMed]
- Jia S, Wildner H, Birchmeier C. Insm1 controls the differentiation of pulmonary neuroendocrine cells by repressing Hes1. Dev Biol 2015;408:90-8. [Crossref] [PubMed]
- Fujino K, Motooka Y, Hassan WA, et al. Insulinoma-associated protein 1 is a crucial regulator of neuroendocrine differentiation in lung cancer. Am J Pathol 2015;185:3164-77. [Crossref] [PubMed]
- Lan MS, Russell EK, Lu J, et al. IA-1, a new marker for neuroendocrine differentiation in human lung cancer cell lines. Cancer Res 1993;53:4169-71. [PubMed]
- Bhattacharjee A, Richards WG, Staunton J, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A 2001;98:13790-5. [Crossref] [PubMed]
- Garber ME, Troyanskaya OG, Schluens K, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A 2001;98:13784-9. [Crossref] [PubMed]
- Hock H, Hamblen MJ, Rooke HM, et al. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature 2004;431:1002-7. [Crossref] [PubMed]
- Zeng H, Yucel R, Kosan C, et al. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J 2004;23:4116-25. [Crossref] [PubMed]
- Wallis D, Hamblen M, Zhou Y, et al. The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Development 2003;130:221-32. [Crossref] [PubMed]
- Kazanjian A, Wallis D, Au N, et al. Growth factor independence-1 is expressed in primary human neuroendocrine lung carcinomas and mediates the differentiation of murine pulmonary neuroendocrine cells. Cancer Res 2004;64:6874-82. [Crossref] [PubMed]
- Shroyer NF, Wallis D, Venken KJ, et al. Gfi1 functions downstream of Math1 to control intestinal secretory cell subtype allocation and differentiation. Genes Dev 2005;19:2412-7. [Crossref] [PubMed]
- Linnoila RI, Jensen-Taubman S, Kazanjian A, et al. Loss of GFI1 impairs pulmonary neuroendorine cell proliferation, but the neuroendocrine phenotype has limited impact on post-naphthalene airway repair. Lab Invest 2007;87:336-44. [Crossref] [PubMed]
- Saleque S, Kim J, Rooke HM, et al. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol Cell 2007;27:562-72. [Crossref] [PubMed]
- Mohammad HP, Smitheman KN, Kamat CD, et al. A DNA hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer Cell 2015;28:57-69. [Crossref] [PubMed]
- Takagi S, Ishikawa Y, Mizutani A, et al. LSD1 inhibitor T-3775440 inhibits SCLC cell proliferation by disrupting LSD1 interactions with SNAG domain proteins INSM1 and GFI1B. Cancer Res 2017;77:4652-62. [Crossref] [PubMed]
- Ntziachristos P, Lim JS, Sage J, et al. From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell 2014;25:318-34. [Crossref] [PubMed]
- Ito T, Udaka N, Yazawa T, et al. Basic helix-loop-helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development 2000;127:3913-21. [PubMed]
- Falix FA, Aronson DC, Lamers WH, et al. Possible roles of DLK1 in the Notch pathway during development and disease. Biochim Biophys Acta 2012;1822:988-95. [Crossref] [PubMed]
- Sriuranpong V, Borges MW, Ravi RK, et al. Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer Res 2001;61:3200-5. [PubMed]
- Lim JS, Ibaseta A, Fischer MM, et al. Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature 2017;545:360-4. [Crossref] [PubMed]
- Ball DW. Achaete-scute homolog-1 and Notch in lung neuroendocrine development and cancer. Cancer Lett 2004;204:159-69. [Crossref] [PubMed]
- Calbo J, van Montfort E, Proost N, et al. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell 2011;19:244-56. [Crossref] [PubMed]
- Oh S, Shin S, Janknecht R. ETV1, 4 and 5: an oncogenic subfamily of ETS transcription factors. Biochim Biophys Acta 2012;1826:1-12. [PubMed]
- Horiuchi S, Yamamoto H, Min Y, et al. Association of ets-related transcriptional factor E1AF expression with tumour progression and overexpression of MMP-1 and matrilysin in human colorectal cancer. J Pathol 2003;200:568-76. [Crossref] [PubMed]
- Sloan KA, Marquez HA, Li J, et al. Increased PEA3/E1AF and decreased Net/Elk-3, both ETS proteins, characterize human NSCLC progression and regulate caveolin-1 transcription in Calu-1 and NCI-H23 NSCLC cell lines. Carcinogenesis 2009;30:1433-42. [Crossref] [PubMed]
- Yuen HF, Chan YK, Grills C, et al. Polyomavirus enhancer activator 3 protein promotes breast cancer metastatic progression through Snail-induced epithelial-mesenchymal transition. J Pathol 2011;224:78-89. [Crossref] [PubMed]
- Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol 2013;14:416-29. [Crossref] [PubMed]
- Watkins DN, Berman DM, Burkholder SG, et al. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature 2003;422:313-7. [Crossref] [PubMed]
- Park KS, Martelotto LG, Peifer M, et al. A crucial requirement for Hedgehog signaling in small cell lung cancer. Nat Med 2011;17:1504-8. [Crossref] [PubMed]
- Szczepny A, Rogers S, Jayasekara WSN, et al. The role of canonical and non-canonical Hedgehog signaling in tumor progression in a mouse model of small cell lung cancer. Oncogene 2017;36:5544-50. [Crossref] [PubMed]
- Yauch RL, Gould SE, Scales SJ, et al. A paracrine requirement for hedgehog signalling in cancer. Nature 2008;455:406-10. [Crossref] [PubMed]
- Pietanza MC, Litvak AM, Varghese AM, et al. A phase I trial of the Hedgehog inhibitor, sonidegib (LDE225), in combination with etoposide and cisplatin for the initial treatment of extensive stage small cell lung cancer. Lung Cancer 2016;99:23-30. [Crossref] [PubMed]
- Chaudhry AZ, Lyons GE, Gronostajski RM. Expression patterns of the four nuclear factor I genes during mouse embryogenesis indicate a potential role in development. Dev Dyn 1997;208:313-25. [Crossref] [PubMed]
- Grabowska MM, Elliott AD, DeGraff DJ, et al. NFI transcription factors interact with FOXA1 to regulate prostate-specific gene expression. Mol Endocrinol 2014;28:949-64. [Crossref] [PubMed]
- Steele-Perkins G, Plachez C, Butz KG, et al. The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol Cell Biol 2005;25:685-98. [Crossref] [PubMed]
- Becker-Santos DD, Lonergan KM, Gronostajski RM, et al. Nuclear factor I/B: a master regulator of cell differentiation with paradoxical roles in cancer. EBioMedicine 2017;22:2-9. [Crossref] [PubMed]
- Dooley AL, Winslow MM, Chiang DY, et al. Nuclear factor I/B is an oncogene in small cell lung cancer. Genes Dev 2011;25:1470-5. [Crossref] [PubMed]
- Semenova EA, Kwon MC, Monkhorst K, et al. Transcription factor NFIB is a driver of small cell lung cancer progression in mice and marks metastatic disease in patients. Cell Rep 2016;16:631-43. [Crossref] [PubMed]
- Denny SK, Yang D, Chuang CH, et al. Nfib Promotes Metastasis through a Widespread Increase in Chromatin Accessibility. Cell 2016;166:328-42. [Crossref] [PubMed]
- Wu N, Jia D, Ibrahim AH, et al. NFIB overexpression cooperates with Rb/p53 deletion to promote small cell lung cancer. Oncotarget 2016;7:57514-24. [PubMed]
- Kirmizis A, Bartley SM, Kuzmichev A, et al. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev 2004;18:1592-605. [Crossref] [PubMed]
- Bracken AP, Dietrich N, Pasini D, et al. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 2006;20:1123-36. [Crossref] [PubMed]
- Arisan S, Buyuktuncer ED, Palavan-Unsal N, et al. Increased expression of EZH2, a polycomb group protein, in bladder carcinoma. Urol Int 2005;75:252-7. [Crossref] [PubMed]
- Bryant RJ, Cross NA, Eaton CL, et al. EZH2 promotes proliferation and invasiveness of prostate cancer cells. Prostate 2007;67:547-56. [Crossref] [PubMed]
- Coe BP, Thu KL, Aviel-Ronen S, et al. Genomic deregulation of the E2F/Rb pathway leads to activation of the oncogene EZH2 in small cell lung cancer. PLoS One 2013;8:e71670. [Crossref] [PubMed]
- Ramaglia M, D'Angelo V, Iannotta A, et al. High EZH2 expression is correlated to metastatic disease in pediatric soft tissue sarcomas. Cancer Cell Int 2016;16:59. [Crossref] [PubMed]
- Zhang H, Qi J, Reyes JM, et al. Oncogenic deregulation of EZH2 as an opportunity for targeted therapy in lung cancer. Cancer Discov 2016;6:1006-21. [Crossref] [PubMed]
- Serresi M, Gargiulo G, Proost N, et al. Polycomb repressive complex 2 is a barrier to kras-driven inflammation and epithelial-mesenchymal transition in non-small-cell lung cancer. Cancer Cell 2016;29:17-31. [Crossref] [PubMed]
- Sato T, Kaneda A, Tsuji S, et al. PRC2 overexpression and PRC2-target gene repression relating to poorer prognosis in small cell lung cancer. Sci Rep 2013;3:1911. [Crossref] [PubMed]
- Hubaux R, Thu KL, Coe BP, et al. EZH2 promotes E2F-driven SCLC tumorigenesis through modulation of apoptosis and cell-cycle regulation. J Thorac Oncol 2013;8:1102-6. [Crossref] [PubMed]
- Gao YB, Chen ZL, Li JG, et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet 2014;46:1097-102. [Crossref] [PubMed]
- Fillmore CM, Xu C, Desai PT, et al. EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature 2015;520:239-42. [Crossref] [PubMed]
- Poirier JT, Gardner EE, Connis N, et al. DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene 2015;34:5869-78. [Crossref] [PubMed]
- Barretina J, Caponigro G, Stransky N, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483:603-7. [Crossref] [PubMed]
- Zoppoli G, Regairaz M, Leo E, et al. Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc Natl Acad Sci U S A 2012;109:15030-5. [Crossref] [PubMed]
- Mu Y, Lou J, Srivastava M, et al. SLFN11 inhibits checkpoint maintenance and homologous recombination repair. EMBO Rep 2016;17:94-109. [Crossref] [PubMed]
- Tang SW, Bilke S, Cao L, et al. SLFN11 is a transcriptional target of ews-fli1 and a determinant of drug response in ewing sarcoma. Clin Cancer Res 2015;21:4184-93. [Crossref] [PubMed]
- Gardner EE, Lok BH, Schneeberger VE, et al. Chemosensitive relapse in small cell lung cancer proceeds through an ezh2-slfn11 axis. Cancer Cell 2017;31:286-99. [Crossref] [PubMed]
- Lee MG, Villa R, Trojer P, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 2007;318:447-50. [Crossref] [PubMed]
- Gherardi S, Ripoche D, Mikaelian I, et al. Menin regulates Inhbb expression through an Akt/Ezh2-mediated H3K27 histone modification. Biochim Biophys Acta 2017;1860:427-37. [Crossref] [PubMed]
- Cimino G, Moir DT, Canaani O, et al. Cloning of ALL-1, the locus involved in leukemias with the t(4;11)(q21;q23), t(9;11)(p22;q23), and t(11;19)(q23;p13) chromosome translocations. Cancer Res 1991;51:6712-4. [PubMed]
- Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell 1992;71:691-700. [Crossref] [PubMed]
- Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333-9. [Crossref] [PubMed]
- Pleasance ED, Stephens PJ, O'Meara S, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 2010;463:184-90. [Crossref] [PubMed]
- Augert A, Zhang Q, Bates B, et al. Small cell lung cancer exhibits frequent inactivating mutations in the histone methyltransferase KMT2D/MLL2: CALGB 151111 (Alliance). J Thorac Oncol 2017;12:704-13. [Crossref] [PubMed]
- Jiang L, Huang J, Higgs BW, et al. Genomic landscape survey identifies SRSF1 as a key oncodriver in small cell lung cancer. PLoS Genet 2016;12:e1005895. [Crossref] [PubMed]
- Metzger E, Wissmann M, Yin N, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 2005;437:436-9. [Crossref] [PubMed]
- Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol 2007;8:307-18. [Crossref] [PubMed]
- Wang J, Scully K, Zhu X, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 2007;446:882-7. [Crossref] [PubMed]
- Kahl P, Gullotti L, Heukamp LC, et al. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res 2006;66:11341-7. [Crossref] [PubMed]
- Lim S, Janzer A, Becker A, et al. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis 2010;31:512-20. [Crossref] [PubMed]
- Lv T, Yuan D, Miao X, et al. Over-expression of LSD1 promotes proliferation, migration and invasion in non-small cell lung cancer. PLoS One 2012;7:e35065. [Crossref] [PubMed]
- Lynch JT, Harris WJ, Somervaille TC. LSD1 inhibition: a therapeutic strategy in cancer? Expert Opin Ther Targets 2012;16:1239-49. [Crossref] [PubMed]
- Serce N, Gnatzy A, Steiner S, et al. Elevated expression of LSD1 (Lysine-specific demethylase 1) during tumour progression from pre-invasive to invasive ductal carcinoma of the breast. BMC Clin Pathol 2012;12:13. [Crossref] [PubMed]
- Wang X, Zhang Y, Nilsson CL, et al. Association of chromosome 19 to lung cancer genotypes and phenotypes. Cancer Metastasis Rev 2015;34:217-26. [Crossref] [PubMed]
- Amente S, Bertoni A, Morano A, et al. LSD1-mediated demethylation of histone H3 lysine 4 triggers Myc-induced transcription. Oncogene 2010;29:3691-702. [Crossref] [PubMed]
- Martinez-Balbás MA, Bannister AJ, Martin K, et al. The acetyltransferase activity of CBP stimulates transcription. EMBO J 1998;17:2886-93. [Crossref] [PubMed]
- Guelman S, Kozuka K, Mao Y, et al. The double-histone-acetyltransferase complex ATAC is essential for mammalian development. Mol Cell Biol 2009;29:1176-88. [Crossref] [PubMed]
- Francetic T, Le May M, Hamed M, et al. Regulation of Myf5 early enhancer by histone acetyltransferase p300 during stem cell differentiation. Mol Biol 2012;1:5019. [PubMed]
- Borrow J, Stanton VP Jr, Andresen JM, et al. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet 1996;14:33-41. [Crossref] [PubMed]
- Giles RH, Petrij F, Dauwerse HG, et al. Construction of a 1.2-Mb contig surrounding, and molecular analysis of, the human CREB-binding protein (CBP/CREBBP) gene on chromosome 16p13.3. Genomics 1997;42:96-114. [Crossref] [PubMed]
- Kasper LH, Qu C, Obenauer JC, et al. Genome-wide and single-cell analyses reveal a context dependent relationship between CBP recruitment and gene expression. Nucleic Acids Res 2014;42:11363-82. [Crossref] [PubMed]
- Choi HJ, Park JH, Park M, et al. UTX inhibits EMT-induced breast CSC properties by epigenetic repression of EMT genes in cooperation with LSD1 and HDAC1. EMBO Rep 2015;16:1288-98. [Crossref] [PubMed]
- Wang XQ, Dostie J. Reciprocal regulation of chromatin state and architecture by HOTAIRM1 contributes to temporal collinear HOXA gene activation. Nucleic Acids Res 2017;45:1091-104. [PubMed]
- Ogiwara H, Sasaki M, Mitachi T, et al. Targeting p300 addiction in CBP-deficient cancers causes synthetic lethality by apoptotic cell death due to abrogation of MYC expression. Cancer Discov 2016;6:430-45. [Crossref] [PubMed]
- Bitler BG, Aird KM, Garipov A, et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med 2015;21:231-8. [Crossref] [PubMed]
- Byers LA, Wang J, Nilsson MB, et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov 2012;2:798-811. [Crossref] [PubMed]
- Romero OA, Torres-Diz M, Pros E, et al. MAX inactivation in small cell lung cancer disrupts MYC-SWI/SNF programs and is synthetic lethal with BRG1. Cancer Discov 2014;4:292-303. [Crossref] [PubMed]
- Jahchan NS, Lim JS, Bola B, et al. Identification and targeting of long-term tumor-propagating cells in small cell lung cancer. Cell Rep 2016;16:644-56. [Crossref] [PubMed]
- Kaur G, Reinhart RA, Monks A, et al. Bromodomain and hedgehog pathway targets in small cell lung cancer. Cancer Lett 2016;371:225-39. [Crossref] [PubMed]
- Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer 2012;12:465-77. [Crossref] [PubMed]
- Lenhart R, Kirov S, Desilva H, et al. Sensitivity of small cell lung cancer to BET inhibition is mediated by regulation of ASCL1 gene expression. Mol Cancer Ther 2015;14:2167-74. [Crossref] [PubMed]
- Kato F, Fiorentino FP, Alibes A, et al. MYCL is a target of a BET bromodomain inhibitor, JQ1, on growth suppression efficacy in small cell lung cancer cells. Oncotarget 2016;7:77378-88. [Crossref] [PubMed]


