
Silence of S1 RNA binding domain 1 represses cell growth and promotes apoptosis in human non-small cell lung cancer cells
Introduction
Lung cancer is the leading cause of cancer death worldwide, and 75–85% of newly diagnosed lung cancers are non-small cell lung cancer with 5-year survival rates approximately 10% (1). Smoking and the use of tobacco products are the main causes of lung cancer cases. Besides, asbestos, air pollution exposures, radon gas, and inherited and acquired mechanisms, also contribute to lung carcinogenesis (2). Today, there are several treatment options for lung cancer, including surgery, radiation therapy, chemotherapy, and targeted therapy. And surgery is the most effective treatment for early-stage or advanced lung cancer, and combination chemotherapy is the standard adjuvant approach, such as platinum-based combined chemotherapy (2). Despite the improvements in diagnosis and therapy of lung cancer, the prognosis for patients with lung cancer is still unsatisfactory.
Lung carcinogenesis is a complex and multistep process, involving the activation of oncogenes and inactivation of tumor suppressor genes. Several reports have identified that several targetable genetic alterations in lung cancer, including activating mutations and amplification of proto-oncogenes, structural rearrangements in ALK and ROS1, oncogenic gene overexpression of miRNA, inactivation of tumor suppressor genes and enhanced telomerase activity (3,4). Although tremendous work has been conducted to translate the findings to improve the patient care in the clinical therapy and diagnosis, survival rates for lung cancer remain dismal (5-8). More studies are still needed to identify the underlying mechanism affecting clinical outcome.
S1 RNA Binding Domain 1 (SRBD1), located at chromosome 2p21, is an RNA binding protein, and initially identified as an S1 RNA-binding domain in Escherichia coli (9). SRBD1 can participate in the regulation of RNA transcription, folding and translation, and indirectly involved in cell growth, general protein synthesis, induction of apoptosis, and maintaining homeostasis (9). Till now, SRBD1 has widely been reported to be susceptibility gene for early-onset normal-tension glaucoma (10-12). Enhanced expression of SRBD1 can lead to increased activity of SRBD1, induce apoptosis, and result in retinal ganglion cell death during the development of glaucoma (10). However, the reports of functions of SRBD1 in other fields were few, including lung carcinogenesis.
In this study, we found that SRBD1 were specifically expressed in the non-small cell lung cancer tissue compared with respective non-cancerous lung tissue. Silencing of SRBD1 inhibited cell proliferation and promoted cell apoptosis in vitro, and suppressed tumorigenesis in a nude mouse model in vivo. Further, we found that SRBD1 silencing reduced the expression of PYCR1, EPS15, HNRNPA0 and IGF1R, and regulated the growth and apoptosis.
Methods
Cell culture and tissue sample collection
Human lung cancer cell lines, A549, NCI-H1299, NCI-H1975 and 95-D cells, were maintained in Ham’s F12 medium (F12) (Corning, USA) supplemented with 10% fetal bovine serum (FBS, Ausbian, China). Cells were incubated at 37 °C in 5% CO2, and the medium was changed every two days. All cell lines were purchased from American Type Culture Collection (ATCC). Eighteen human lung cancer tissues, adjacent normal lung tissue (NAT) and cancer adjacent lung tissue (AT) were obtained from lung cancer patients with informed consent and Institutional Review Board permission. Samples were separated from the surgical patient samples, and immediately snap-frozen and stored in liquid nitrogen until use.
Synthesis and transfection of shRNA
shRNAs were designed and synthesized by Genechem group (Shanghai, China). The most effective siRNA, SRBD1 directed-shRNA (shSRBD1) was identified by qPCR and applied for the further experiments. The interference sequence was GCCAACATCATTCGTCTCTTTCTCGAGAAAGAGACGAATGATGTTGGC. A549 and HCI-H1299 cells were digested into single cells by trypsin (Sangon Biotech, China) and seeded in 6-well plate culture. When the cell density grew to 2×105/well, the cells were randomly divided into control and experimental groups for transfection. After 12 h, the new complete medium was changed, and expression of green fluorescent protein (GFP) was detected by fluorescence microscope (Olympus, Japan) 24, 48 and 72 h after transfection. When the transfection efficiency was over 80%, these cells were used to detect the effects of shSRBD1 on A549 and HCI-H1299 cells.
Proliferation assay
When transfected A549 and NCI-H1299 cells grew to 80% confluence, the cells were digested into single cell suspension by trypsin, and seeded into a 96-well plate in cell density of 2×103/well. Celigo (Nexcelom, USA) were used to read the plate for 3–5 days and analyze the curve of cell proliferation. And 20 µL 5 mg/mL MTT solution (Genview, USA) was added into the well on day 1 to 5, and the cells were incubated at 37 °C for 4 h. Then, the culture medium was abandoned, and 100 µL/well dimethyl sulfoxide (Shiyicr company, China) was added. OD value was detected by microplate reader (Tecan infinite, Switzerland) at 490/570 nm.
Clone formation was detected by crystal violet staining. The transfected cells were seeded into 6-well cell culture plate in 1×103/well. After 15 days’ culture, the cells were stained by crystal violet solution (Sangon Biotech, China), and the clone were photographed by digital camera, and counted.
Apoptosis assay
When transfected A549 and NCI-H1299 cells grew to 80% confluence, the cells in suspension were collected, and the cells on the dish were digested into single cells suspension by trypsin. All cells were collected for apoptosis assay Annexin V-APC apoptosis detection kit (eBioscience, USA) according to the manufacturer’s protocol and FACS analysis (Millipore, USA).
Microarray analysis
Total RNA of control-shRNA (shCtrl) and shSRBD1 transfected A549 cells were isolated using RNA 6000 Nano Kit (Agilent Technologies, USA). The microarray platform in this study was Genechip PrimeView TM human (Affymetrix, USA). The gene expression profile was preprocessed using Limma (13) (version 3.83, linear models for microarray data, www.bioconductor.org/packages/2.8/bioc/html/limma.html) package in Bioconductor Affymetrix annotation files from Brain Array Lab (version 20, http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/CDF_download.asp). The normalized data were calculated with the Limma package, and genes with P<0.05 and |log2fold change| ≥1.5 were considered to indicate a statistically significant difference between the group and the group. Ingenuity Pathway Analysis of differentially expressed genes was conducted with Qiagen’s Ingenuity Pathway Analysis algorithm (www.qiagen.com/ingenuity, Qiagen, Redwood City, CA, USA). Canonical pathway analysis, upstream analysis and interaction network analysis were performed.
Tumor formation in BALB/c nude mice
All experimental procedures involving animals were performed according to the institutional ethical guidelines for animal experiments at the Peking Union Medical College. A549 cells were transfected with shCtrl and shSRBD1, respectively. 1×107/200 µL tranfected A549 cells were transplanted into BALB/c athymic nude mouse (4 weeks of age, n=10) by hypodermic injection in oxter. The growth of tumors was examined every day. Tumor volume (V) was monitored by measuring the length (L) and width (W) with calipers, and calculated with the formula V= (L×W2)×π/6. Seven days after cell injection, D-luciferin (15 mg/mL, Qcbio, China) was intraperitoneally injected at a dose of 10 µL/g, and live images of the mouse were obtained using in vivo imaging system (Perkin Elimer, Germany). Tumor weight was measured by tray balance (Sartorius, Germany).
Western blot analysis
shCtrl and shSRBD1 transfected A549 cells were lysed in RIPA buffer (Beyotime, China) containing EDTA-free protease inhibitor cocktail (Roche, USA). Protein samples were separated via 6–10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes (Millipore, Billerica, USA). Membranes were blocked in 5% bovine serum albumin for 1 h and then incubated with primary antibodies (Table 1) overnight at 4 °C. Horseradish peroxidase (HRP)-conjugated Monoclonal Mouse Anti-GAPDH (Kangchen, Shanghai, China) was used as the internal control. After treatment with HRP secondary anti-mouse or anti-rabbit antibodies (Santa Cruz, USA), imaging was performed using medical X-ray film (Carestream, Canada).
Full table
RNA isolation and RT-PCR
Total RNA was extracted from shCtrl and shSRBD1 transfected A549 cells by trizol (Pufei, Shanghai, China). cDNAs were synthesized using M-MLV-reverse transcriptase kit (Promega, USA) according to the manufacturer’s protocol. QPCR was carried out using SYBR® Green reagents and the Applied Biosystems 7500 Real Time PCR System (Applied Biosystems, Foster City, CA) and the data were analyzed with the Sequence Detection System software (Applied Biosystems) using GAPDH as an internal control (Table 2).
Full table
Statistical analysis
Data are representative of at least 3 independent experiments, and presented as means ± SD in this study. Statistical analysis was performed using SPSS 17.0 software (SPSS, Chicago, USA). The differences were tested with Student’s t-test. A P<0.05 was considered to be statistically significant.
Results
SRBD1 was highly expressed in squamous lung cancer tissue
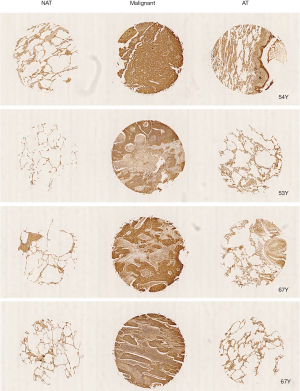
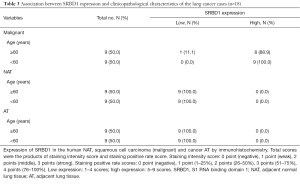
To explore whether SRBD1 was expressed in human non-small cell lung cancer cells, the expression of SRBD1 protein in human NAT, squamous cell carcinoma (malignant) and cancer AT was analyzed by immunohistochemistry and in situ hybridization. As shown in Figure 1, all malignant tissues highly expressed SRBD1, while expressions of SRBD1 in NAT and AT tissues were low. Besides, SRBD1 staining was quantified by scores, which are the products of staining intensity score and staining positive rate score. Consistently, scores of SRBD1 expressions in malignant tissues were all high, except one case in the ≥60 group. However, SRBD1 expressions in NAT and AT tissues absolutely exhibited low scores (Table 3). These data suggested SRBD1 was specifically expressed in human squamous cell carcinoma.
Full table
Expression of SRBD1 was silenced by shRNA in human non-small cell lung cancer cells
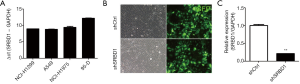
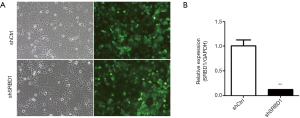
The expression levels of SRBD1 mRNA in 4 lung cancer cell lines, NCI-H1299, A549, NCI-H1975 and 95-D, were analyzed by RT-PCR. Normalization was performed using GAPDH as an internal control. Except 95-D, the lung cancer cell lines, NCI-H1299, A549 and NCI-H1975, expressed high levels of SRBD1 mRNA (Figure 2A). Then, A549 and NCI-H1299 were selected to detect the effect of SRBD1 in lung cancer cells. A549 cells were firstly transfected with lentivirus shCtrl and shSRBD1, respectively. As shown in Figure 2B, transfection efficiencies were estimated by statistics of GFP positive cells, and the percentages of GFP + cells were over 85% 72 h later. Moreover, the expressions of SRBD1 mRNA with shCtrl or shSRBD1 transfection were detected by RT-PCR. Compared with the shCtrl group, the expression of SRBD1 in shSRBD1 group reduced to 20% of the control (P<0.01) (Figure 2C). Besides, NCI-H1299 was also transfected with shCtrl and shSRBD1, respectively, and showed the similar trend (Figure S1).
SRBD1 silencing inhibited cell proliferation of human non-small cell lung cancer cells
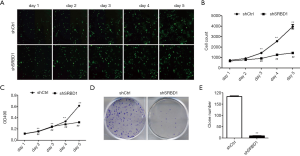
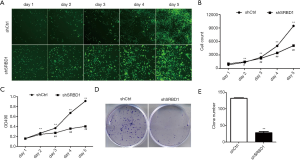
To further evaluate the biological effects of SRBD1 silencing in human non-small cell lung cancer cell, cell proliferation was assessed using cell counting, MTT assay and clone formation. As shown in Figure 3A,B, the suppression of SRBD1 had a direct effect on cell proliferation. The cell number of shCtr-treated cells showed a upward trend in 5 days’ culture, however, the change of the number of shSRBD1 group was not significant. MTT assay was used to detected cell viability. Compared the upward trend in shCtrl group, the growth of OD490 absorbance value in shSRBD1 group was slow (Figure 3C). Besides, the ability of clone formation was also detected. In Figure 3D, shCtrl-A549 formed large and dense cell clones, while shSRBD1-A549 exhibited small and few clones. Statistical analysis also showed that the number of clones in shSRBD1 group was significantly lower than ones in the control group (P<0.01) (Figure 3E). These results were also confirmed in shSRBD1-treated NCI-H1299 cells (Figure S2), suggesting that the silencing of SRBD1 expression inhibited the proliferation of human non-small cell lung cancer cells.
SRBD1 silencing promoted cell apoptosis of human non-small cell lung cancer cells
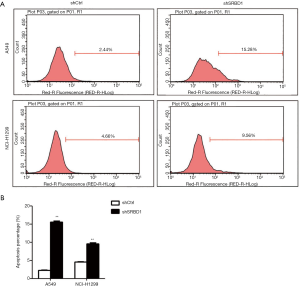
To detect the effect of SRBD1 silencing on the apoptosis of non-small cell lung cancer cells, we performed Annexin V-APC flow analysis. The results showed that the apoptosis rate of shSRBD1 treated A549 cells was significantly higher than the control group (15.61%±0.44% vs. 2.23%±0.19%) (P<0.01). Similar results were also got in shSRBD1 treated NCI-H1299 cells (Figure 4). These results suggest that silencing of SRBD1 expression can promote the apoptosis of human non-small cell lung cancer cells.
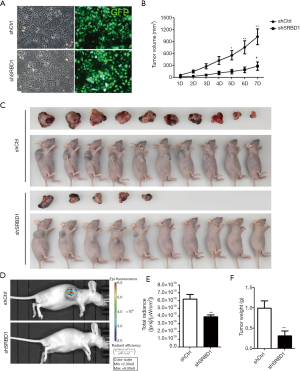
SRBD1 silencing inhibited tumorigenicity of human non-small cell lung cancer cells in vivo
The above results demonstrated that SRBD1 silencing inhibited cell growth and promoted cell apoptosis of human non-small cell lung cancer cells. To detect whether SRBD1 silencing inhibited tumorigenicity in vivo, A549 cells were transfected by shCtrl or shSRBD1 (Figure 5A), and 1×107 cell suspension of the transfected cells were transplanted into BALB/c nude mice xenograft model by hypodermic injection in oxter. Tumor volume were measured every day until 7 days after transplantation. Tumor volume in the shSRBD1-A549 injected group was significantly smaller than the volume in the shCtrl group from day 1 (P<0.01) (Figure 5B). Different from the big tumor formed in the mice with shCtrl A549 injection, there were only small or no tumor formation in the mice with shSRBD1-A549 injection 7 days after treatment (Figure 5C). The luciferase activity was also detected on day 7. Fluorescence intensity of shSRBD1-A549 cells were significantly lower than the control group (P<0.01) (Figure 5D,E). Besides, tumor weights isolated from these two groups were measured, and the weights of the shCtrl group were dramatically higher than the weights of shSRBD1 group (P<0.01) (Figure 5F). These results suggested that silencing of SRBD1 expression could inhibit the tumorigenicity of human non-small cell lung cancer cells in vivo.
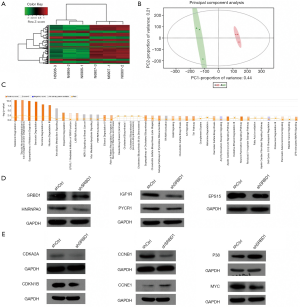
SRBD1 promoted proliferation and resisted apoptosis through ELAVL1 -mediated IGF1R/HNRNPA0 and NTRK1-mediated EPS15/PYCR1
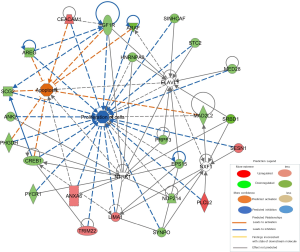
To explain the mechanism of SRBD1 functions in human non-small cell lung cancer cells, microarray analysis and principal component analysis of differentially expressed genes in A549 cells with shCtrl or shSRBD1 transfection showed that silencing of SRBD1 expression resulted in marked changes in gene expression (Figure 6A,B). Classical pathway analysis showed that several pathways were significantly activated in the shSRBD1 treated A549 cells, including melatonin degradation, thyroid hormone metabolism, serotonin degradation, nicotine degradation and dopamine degradation (Figure 6C). Besides, IPA was used to further explore potential signaling pathways. We screened the disease/function-related and cancer cell proliferation-related genes from differentially expressed genes, and showed the predicted molecular regulatory network of SRBD1 silencing inhibiting tumor cell proliferation and promoting apoptosis, which shows the network of interactions and regulatory relationships between SRBD1 and molecules involved in cell proliferation and apoptosis (Figure S3). Then, the expression of all these factors in mRNA level were firstly detected by RT-PCR, and expressions of 21 factors were significantly downregulated in shSRBD1 group (Figure S4). Meanwhile, we also detected the protein level of all these factors by western blot. The results showed that the protein levels of the apoptosis-related factor, IGF1R, and the proliferation-related factors, EPS 15, PYCR1 and HNRNPA0, were downregulated in shSRBD1 group (Figure 6D). In addition, the expressions of the classical factors involved in the growth and apoptosis of cancer cells were also analyzed by western blot. We found that, compared with the expression of the shCtrl group, the expressions of cell-cycle control genes, CDKN2A, CDKN1B, CCNB1 and CCNE1, and the expression of the oncogene, Myc, significantly decreased in shSRBD1 transfected A549 cells. P38, one major signaling molecule, increased in shSRBD1 group (Figure 6E). These results suggest that SRBD1 promotes proliferation and resists apoptosis via IGF1R/EPS15/PYCR1/HNRNPA0.

Discussion
It has been well documented that SRBD1 exhibited significant association with glaucoma (11,14). Previous studies also reported that SRBD1 was involved in cell growth and apoptosis (10). However, the detailed effect of SRBD1 and its functional mechanism in non-small cell lung cancer remain unclear. In the current study, for the first time, we demonstrated that SRBD1 was specifically expressed in human non-small cell lung cancer tissue. Further study revealed that SRBD1 silencing inhibited cell proliferation and promoted cell apoptosis of A549 and NCI-H1299 cells, and suppressed tumorigenesis of non-small cell lung cancer cells in vivo. In addition, our results further revealed SRBD1 silencing played the roles via P53 and JNK pathways by reducing the expressions of EPS 15, IGF1R, MYC, PYCR1 and HNRNPA0.
Abnormal cell proliferation and apoptosis plays an important role in the occurrence of tumors. Different from normal cells, the death and apoptosis of malignant cell had defects, which caused tumor cells to grow out of control. Most chemotherapeutic agents were designed to kill tumor cells by inducing apoptosis, such as mapatumumab, apomab, conatumumab and YM155 (15-18). Recently, to inhibit the growth and promote apoptosis of non-small cell lung cancer cells by genetic technology attracts more and more attentions. In this study, eighteen groups of human lung NAT, malignant and AT were used to detected SRBD1 expression. For the first time, we found SRBD1 was specifically expressed in human squamous cell carcinoma, which suggested SRBD1 might be a new diagnostic indicator of non-small cell lung cancer. To explore the functions of SRBD1 in human non-small cell lung cancer, we silenced the expression of SRBD1 of A549 and NCI-H1299 cells by shRNA. The results showed silencing of SRBD1 could significantly inhibit cell growth and promote apoptosis in A549 and NCI-H1299 cells, and effectively inhibit tumorigenicity BALB/c nude mice xenograft model, so silencing the expression level of SRBD1 can be as a new strategy of treatment of lung adenocarcinoma.
In order to understand the mechanism of SRBD1 silencing affecting the biological characteristics of cancer cells, we used microarray to analyze the shSRBD1 transfected A549 cells. Silencing of SRBD1 expression resulted in marked changes in gene expression of A549 cells. Previous studies have reported that SRBD1 can directly interact with neurotrophic tyrosine kinase 1 (NTRK1) (19). NTRK1 belongs to the neurotyrosine kinase family, and is found in non-small cell lung cancer, colorectal cancer, intrahepatic bile duct cancer, papillary thyroid cancer, parotid gland tumor, glioma and sarcoma cancer cells (20). Overexpression of NTRK1 can activate ERK, PI3k-akt and MAPK pathways, and promote cell growth, metastasis and invasion (21). As shown in our predicated regulatory network, NTRK1 interacts with PYCR1, EPS15 and HNRNPA0 to regulate proliferation of tumor cells. SRBD1 silencing reduced the expressions of PYCR1, EPS15 and HNRNPA0, and inhibited cell proliferation of lung cancer cells, which suggested SRBD1 might promote proliferation through NTRK1-mediatd PYCR1, EPS15 and HNRNPA0.
SRBD1 was also reported to interact with ELAV Like RNA Binding Protein 1 (ELAVL1), a member of the ELAV RNA binding protein family (22). ELAVL1 can selectively bind to the AU enrichment region at the 3'-utrterminal of mRNA. By targeting the mRNA of downstream genes, ELAVL1 can promote cell proliferation, survival and angiogenesis, and promote the invasion and metastasis of cancer cells (23). In addition, ELAVL1 can directly interact with IGF1R (24). IGF can bind to IGF1R, lead to tyrosine auto-phosphorylation of IGF1R receptor β subunits, and activate the downstream PI3K/AKT pathway and Ras/MAPK pathway, stimulating cell proliferation and inhibiting apoptosis (25,26). Here, expression of IGF1R decreased in the shSRBD1-treated A549 cells. All these suggests SRBD1 functions through the interaction with ELAVL1-mediated IGF1R in human non-small cell lung cancer cells. The detailed mechanism of SRBD1 silencing affecting the non-small cell lung cancer cells needs the further experiments.
Anomalies in cell-cycle control genes have frequently been observed in many human malignancies, such as breast cancer, ovarian cancer and lung cancer (27-29). Here, we found silencing of SRBD1 reduced the expressions of cell-cycle control genes, CDKN2A, CDKN1B, CCNB1 and CCNE1. Besides, MYC is one of the most frequently deregulated oncogenes in human cancer, and tumors can employ multiple genetic and epigenetic mechanisms to upregulate MYC and commonly become dependent on MYC for proliferation and survival (30). SRBD1 silencing can also effectively reduced the expressions of MYC. In addition, MAPK signaling pathway is an important intracellular signaling system, and activation of the p38 MAPK pathway is essential for drug-induced tumor cell death (31). Expression of p38 in A549 cells was increased after shSRBD1 transfection.
In summary, our results demonstrate the following: SRBD1 is specifically expressed in human non-small cell lung cancer tissues compared to adjacent non-cancerous lung tissues; SRBD1 silencing inhibits cell proliferation and induces cell apoptosis in non-small cell lung cancer cells, and inhibits tumorigenicity in a nude mouse model in vivo; and SRBD1 may promote the proliferation and resist apoptosis through functions through NTRK1-mediatd PYCR1, EPS15 and HNRNPA0 and ELAVL1-mediated IGF1R. Our data suggest that SRBD1 may be an important tumor promoter that could be exploited in non-small cell lung cancer diagnosis and therapy.
Acknowledgments
Funding: This work was supported by CAMS Innovation Fund for Medical Sciences (2017-I2M-1-009).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by institutional ethics committee of Genechem (No. GSGC0156770).
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Lemjabbar-Alaoui H, Hassan OU, Yang YW, et al. Lung cancer: Biology and treatment options. Biochim Biophys Acta 2015;1856:189-210. [PubMed]
- Shtivelman E, Hensing T, Simon GR, et al. Molecular pathways and therapeutic targets in lung cancer. Oncotarget 2014;5:1392-433. [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Asamura H, Chansky K, Crowley J, et al. The International Association for the Study of Lung Cancer Lung Cancer Staging Project: Proposals for the Revision of the N Descriptors in the Forthcoming 8th Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2015;10:1675-84.
- Pereira JL, Gomes M, Teixeira AL, et al. Potential and importance of metalloproteinases and interleukins in inflammation and metastasization in non-small cell lung cancer. Transl Cancer Res 2018;7:782-95. [Crossref]
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Subramanian AR. Structure and functions of ribosomal protein S1. Prog Nucleic Acid Res Mol Biol 1983;28:101-42. [Crossref] [PubMed]
- Kanemaki N, Tchedre KT, Imayasu M, et al. Dogs and humans share a common susceptibility gene SRBD1 for glaucoma risk. PloS one 2013;8:e74372. [Crossref] [PubMed]
- Writing Committee for the Normal Tension Glaucoma Genetic Study Group of Japan Glaucoma Society, Meguro A, Inoko H, et al. Genome-wide association study of normal tension glaucoma: common variants in SRBD1 and ELOVL5 contribute to disease susceptibility. Ophthalmology 2010;117:1331-8.e5. [Crossref] [PubMed]
- Mabuchi F, Sakurada Y, Kashiwagi K, et al. Association between SRBD1 and ELOVL5 gene polymorphisms and primary open-angle glaucoma. Invest Ophthalmol Vis Sci 2011;52:4626-9. [Crossref] [PubMed]
- Diboun I, Wernisch L, Orengo CA, et al. Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics 2006;7:252. [Crossref] [PubMed]
- Mabuchi F, Sakurada Y, Kashiwagi K, et al. Involvement of genetic variants associated with primary open-angle glaucoma in pathogenic mechanisms and family history of glaucoma. Am J Ophthalmol 2015;159:437-44.e2. [Crossref] [PubMed]
- Tolcher AW, Mita M, Meropol NJ, et al. Phase I pharmacokinetic and biologic correlative study of mapatumumab, a fully human monoclonal antibody with agonist activity to tumor necrosis factor-related apoptosis-inducing ligand receptor-1. J Clin Oncol 2007;25:1390-5. [Crossref] [PubMed]
- Jin H, Yang R, Ross J, et al. Cooperation of the agonistic DR5 antibody apomab with chemotherapy to inhibit orthotopic lung tumor growth and improve survival. Clin Cancer Res 2008;14:7733-40. [Crossref] [PubMed]
- Paz-Ares L, Bálint B, de Boer RH, et al. A randomized phase 2 study of paclitaxel and carboplatin with or without conatumumab for first-line treatment of advanced non-small-cell lung cancer. J Thorac Oncol 2013;8:329-37. [Crossref] [PubMed]
- Cheng CC, Chang J, Huang SC, et al. YM155 as an inhibitor of cancer stemness simultaneously inhibits autophosphorylation of epidermal growth factor receptor and G9a-mediated stemness in lung cancer cells. PloS One 2017;12:e0182149. [Crossref] [PubMed]
- Emdal KB, Pedersen AK, Bekker-Jensen DB, et al. Temporal proteomics of NGF-TrkA signaling identifies an inhibitory role for the E3 ligase Cbl-b in neuroblastoma cell differentiation. Sci Signal 2015;8:ra40. [Crossref] [PubMed]
- Farago AF, Le LP, Zheng Z, et al. Durable Clinical Response to Entrectinib in NTRK1-Rearranged Non-Small Cell Lung Cancer. J Thorac Oncol 2015;10:1670-4. [Crossref] [PubMed]
- Cheng Y, Diao DM, Zhang H, et al. Proliferation enhanced by NGF-NTRK1 signaling makes pancreatic cancer cells more sensitive to 2DG-induced apoptosis. Int J Med Sci 2013;10:634-40. [Crossref] [PubMed]
- Abdelmohsen K, Srikantan S, Yang X, et al. Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J 2009;28:1271-82. [Crossref] [PubMed]
- Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip Rev RNA 2010;1:214-29. [Crossref] [PubMed]
- Blume SW, Jackson NL, Frost AR, et al. Northwestern profiling of potential translation-regulatory proteins in human breast epithelial cells and malignant breast tissues: evidence for pathological activation of the IGF1R IRES. Exp Mol Pathol 2010;88:341-52. [Crossref] [PubMed]
- Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell 2010;141:1117-34. [Crossref] [PubMed]
- Logue JS, Morrison DK. Complexity in the signaling network: insights from the use of targeted inhibitors in cancer therapy. Genes Dev 2012;26:641-50. [Crossref] [PubMed]
- Agarwal R, Gonzalez-Angulo AM, Myhre S, et al. Integrative analysis of cyclin protein levels identifies cyclin b1 as a classifier and predictor of outcomes in breast cancer. Clin Cancer Res 2009;15:3654-62. [Crossref] [PubMed]
- Barbieri F, Cagnoli M, Ragni N, et al. Increased cyclin D1 expression is associated with features of malignancy and disease recurrence in ovarian tumors. Clin Cancer Res 1999;5:1837-42. [PubMed]
- Vincenzi B, Schiavon G, Silletta M, et al. Cell cycle alterations and lung cancer. Histol Histopathol 2006;21:423-35. [PubMed]
- Gabay M, Li Y, Felsher DW. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med 2014. [Crossref] [PubMed]
- Feng Y, Liu F, Du Z, et al. Wip1 regulates SKOV3 cell apoptosis through the p38 MAPK signaling pathway. Mol Med Rep 2017;15:3651-7. [Crossref] [PubMed]


